Gene-editing THP-1 cells help reveal the action mechanism of the pro-inflammatory factors CypA
Background
Inflammation is the innate immunity, one of the individual defense mechanisms, a moderate response is beneficial for clearing pathogenic microorganisms and repairing damaged tissues, but excessive inflammation can lead to further tissue damage and even endanger life. Inflammation can be generally divided into three phases, inflammation activation, inflammation resolution, as well as tissue repair and homeostatic remodeling. And the inflammatory pathway is composed of inducers, sensors, mediators and effectors. Once triggered by inducers, inflammatory signals promote the production of inflammatory cytokines such as interleukin-1 (IL-1), tumor necrosis factor-α (TNF-α), IL-6, etc. and participate in signal transduction, playing different roles in different stages of inflammation. Among the IL-1 family, IL-1β is a typical pro-inflammatory cytokine, and its expression is tightly controlled by dual signals. The initial signal can induce the expression of biologically inactive pro-IL-1β, activating signals to activate the inflammasome, and subsequently mature caspase-1 cleaves pro-IL-1β into biologically active IL-1β which will be then secreted outside the cell to function. It has been reported that in the absence of activating signals, the production and secretion of IL-1β are inefficient. Most of the newly synthesized product remains intracellular, whereas this process is regulated by the ubiquitination of pro-IL-1β. Furthermore, IL-1β enables to induce IL-6 and TNF-α production, and by binding to the receptor IL-1RI, IL-1β then recruits the adaptor MyD88/IRAK/TRAF6/NF-kB, forming a signaling cascade.
Cyclophilin A (CypA), a peptidyl
-prolyl cis-trans isomerase encoded by the
PPIA gene, is widely present in various tissues and involved in influenza virus replication, antiviral innate immunity, and bacterial coinfection secondary to influenza virus. It has been reported that CypA is able to promote the expression of inflammatory factors such as IL-1β by regulating RIG-I/MAVS/NF-κB signaling and IL-6 trans
-signaling, but how CypA functions at different stages of inflammation remains unknown. Lei Sun and Wenjun Liu have led their team for systematic research on CypA for more than a decade and deeply researched the CypA in terms of its role and mechanism of action in inflammatory activation, inflammation resolution, and tissue repair processes, and published an article entitled “
Delicate regulation of IL-1β-mediated inflammation by cyclophilin A” on Cell Reports. So now let’s see what materials and methods were used to study the pro-inflammatory factor CypA.

Identification of key cytokines in the inflammatory response from Ppia-/- mouse models
Ppia-/- mice were purchased from Jackson Laboratory and together with wild-type mice (WT) to form lung inflammation mouse models with different time points after LPS induction for 0, 6h, 12h, 24h, 3d, 5d, 7d. First by histopathological staining experiments (Fig. 1A), lung sections from mice at different time points were pathologically scored (Fig. 1B) and, at the same time points, lung dry and wet mass changes (Fig. 1C) and lung index (Fig. 1D) were statistically analyzed and their results were found to be astonishingly similar, thereby illustrating that CypA encoded by Ppia plays different roles at different stages of inflammation.
Because cytokines play an important role in the inflammatory response, to examine how these cytokines behave in the inflammatory models, researchers extracted mRNA from the lungs, bronchoalveolar lavage fluid (BALF), and serum of Ppia-/- and WT mice, respectively, then examined the expression of pulmonary cytokine Il1b (Figure 1E), as well as secretion cytokine IL-1β from BALF (Figure 1F), serum (Figure 1G) and lung (Figures 1H and 1I) by qPCR, ELISA and immunohistochemistry experiments. It was found that the expression of these two cytokines in different parts of WT mice was higher than in Ppia-/- mice at 6h, 12h and 24h, while at 3d, 5d and 7d, the expression of these cytokines was reversed. In addition to this, researchers also examined the expression amount of other cytokines in the various sites (see supplemental figure). Taken together, all experimental findings suggest that in the mouse model of LPS-induced inflammation, CypA acts primarily by regulating IL-1β expression to control its role in different stages of inflammation.
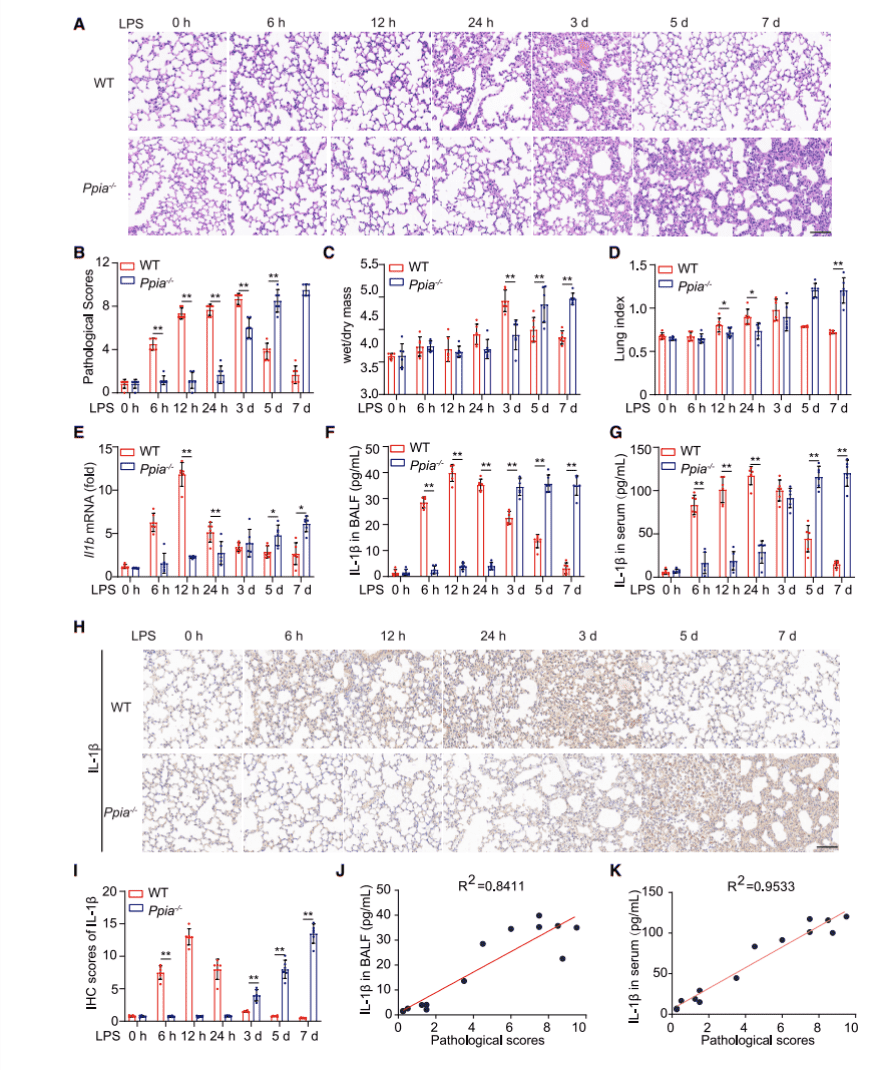
Figure 1. CypA regulates LPS-induced lung inflammation, depending on IL-1b expression
Mechanistic studies of key cytokines in typical lung inflammation cell lines
(1) Promotion of CypA in the early phase of LPS-induced inflammation
To further determine why CypA promotes IL-1β expression during the early phase of inflammation, first through reviewing the literature, it suggests that CypA can regulate p65 from NF-κB to promote proinflammatory cytokine production. Here researchers selected the classical mouse primary cell lines Bone Marrow-Derived Macrophages (BMDM), Human non-small cell lung cancer cell line A549 and Human cell line THP-1 which is a typical cell line for inflammation studies (THP-1/PPIA-/- cell line was provided by Ubigene), and the corresponding mutant cell lines with PPIA knockout or knockdown were generated by CRISPR-Cas9, interference and so on. Subsequently, these WT and mutant cell lines were treated by LPS to mimic lung inflammation, and the amounts of mRNA expression of the cytokines Il1b or IL1B, which have been examined in mouse models, were detected and compared at 0, 2h, 4h, 6h, 8h (Figure S2A-C). Also, through immunoblotting, the amounts of protein expression of CypA, pro-IL-1β at 0, 4h, 8h, and 12h were analyzed as well as the expression amount of phosphorylated p65 at 0, 15min, 30min, 60min (Figure S2G-I). Thus, it was demonstrated that CypA increases the expression of Il1b and IL1B as well as pro-IL-1β by increasing the phosphorylation level of p65. To further determine how CypA promotes IL-1β expression, researchers mutated the functional domain of CypA for PPIase activity (R55A) and examined the same components (Figure S2J-L) as the PPIA knockout cell line, showing consistent results with when the PPIA was knocked out. Thereby illustrating that, in the early phase of inflammation, CypA-mediated enhancement of IL-1β expression requires the activity of its enzyme.
As a typical tool to study immunity and inflammation, THP-1 gene-edited cells, however, often face difficulties of transfection and low monoclonal growth rate, Ubigene through more than 100 real projects, has summarized and performed the targeted optimization of the gene-editing system of THP-1 cell line, which can greatly improve the success rate of gene-editing of THP-1. Click here for more details of our gene-editing cell line services, Ubigene can help you with pathophysiological studies of disease and high throughput drug screening >>
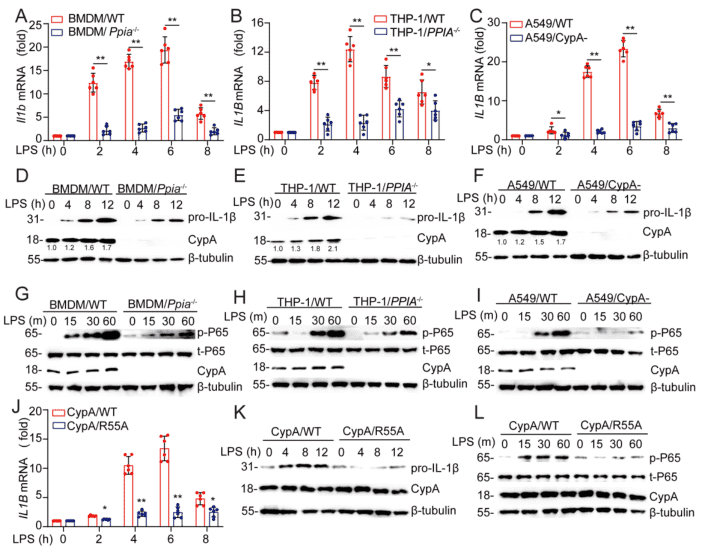
Figure S2. CypA increases pro-IL-1β expression through NF-κb. Related to Figure 2
According to the previous results, IL-1β is initially produced as a cytosolic precursor, an inactive procytokine (pro-IL-1β), and once secondary signals are activated, pro-IL-1β is then cleaved into active IL-1β by the NLRP3/ASC/caspase-1 inflammasome. Therefore, researchers further investigated whether CypA contributes to pro-IL-1β processing. First, through ELISA experiments in BMDM/Ppia-/- (Figure 2A), THP-1/PPIA-/- (Figure S3A), and A549/CypA- (Figure S3B) models, it was observed that when inflammasome activation was stimulated by ATP, IL-1β expression in CypA mutant cell lines was significantly lower than that of the corresponding WT cell line.
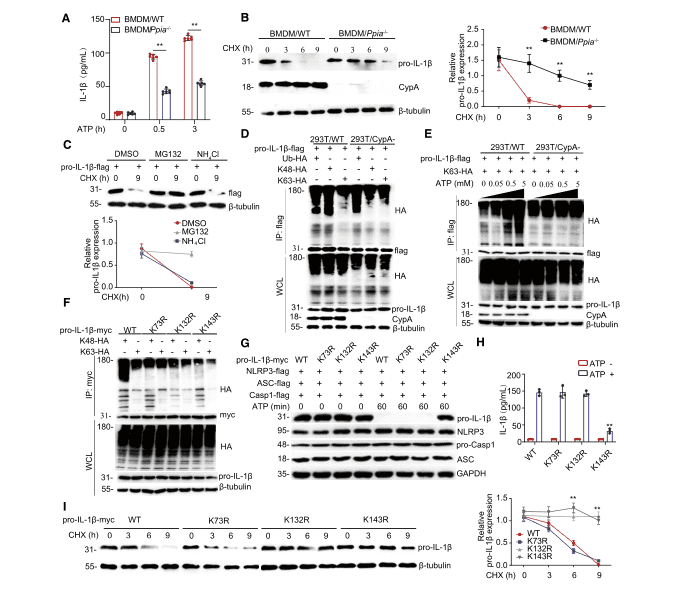
Figure 2. CypA promotes pro-IL-1b processing and degradation by regulating its K63- or K48-linked ubiquitination
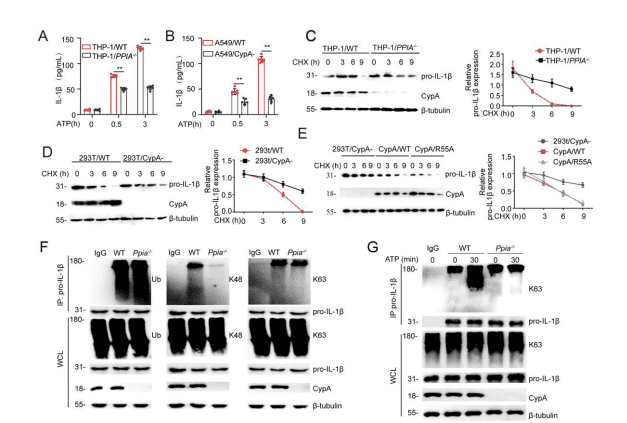
Figure S3. CypA promotes the secretion of IL-1β and pro-IL-1β degradation by regulating proIL-1β ubiquitination. Related to Figure 2.
(2) Inhibition of CypA during the resolution phase of LPS-induced inflammation
It was reported that in the absence of stimulation by secondary signals such as ATP the processing and release of IL-1β are inefficient, and most of the synthesized products are left inside the cell, either left unprocessed or degraded. To explore why CypA inhibits IL-1β expression in the late phase of inflammation, researchers then investigated the effect of CypA on pro-IL-1β stability. First, pro-IL-1β expressions in BMDM/Ppia-/- (Figure 2B), THP-1/Ppia-/- (Figure S3C), 293T/CypA- models were detected using immunoblotting and the results showed that CypA could accelerate the degradation of endogenous pro-IL-1β in the three WT cells described above. Subsequently, a FLAG-tagged pro-IL-1β expression vector was transfected into 293T and the cells were treated with DMSO, MG132, and NH4Cl (Figure 2C) and, for comparison, MG132 was found to promote pro-IL-1β degradation, thereby illustrating that pro-IL-1β degradation is by the ubiquitin-proteasome pathway, and this process is independent of the ppiase activity of CypA (Figure S3E).
(3) CypA affects pro-IL-1β ubiquitination
It has been reported that K63-linked ubiquitination of pro-IL-1β has an important role in the activation of the inflammasome and secretion of IL-1β. Therefore, to verify whether CypA affects the ubiquitination of pro-IL-1β, researchers performed ubiquitination experiments, and by immunoblotting, found in 293T and BMDM cell lines that, in the absence of ATP stimulation, CypA had a significant effect on K48-linked ubiquitination (Figure 2D, FigureS3F). But when ATP was used alone, CypA progressively increased K63-linked ubiquitination of pro-IL-1β (Figure 2E, Figure S3G).
(4) Validation of the key ubiquitination site of pro-IL-1β
To further validate the key ubiquitination site of pro-IL-1β, three potential ubiquitination sites, K73, K132, and K143, were identified by mass spectrometric analysis. Immunoblotting experiments demonstrated that K73R, K132R and K143R mutations resulted in reduced K48-linked pro-IL-1β ubiquitination, whereas the K132R, K143R mutations resulted in reduced K63-linked pro-IL-1β ubiquitination (Figure 2F). At this point, it remained uncertain which the key ubiquitination site for pro-IL-1β processing is. Therefore NLRP3, ASC, and caspase-1 as well as wild-type or mutant forms of pro-IL-1β were co-transfected into 293T cell line which was subsequently stimulated with ATP. The K143R mutation was found to prevent the production of pro-IL-1β to be cleaved into active IL-1β by immunoblot (Figure 2G) and ELISA (Figure 2H) Thus K143 was determined to be the key site for pro-IL-1β processing. Then 293T transfected with vectors encoding wild-type pro-IL-1b-myc and the different mutant forms were tested again by immunoblotting (Figure 2I), and K132 and K143 were found to be the key sites for pro-IL-1β degradation.
Overall, these data indicate that, during the inflammatory activation phase, stimulated by high concentrations of ATP, CypA promotes pro-IL-1β processing mainly by enhancing K63-linked ubiquitination at the K143 site. However, in the remission phase of inflammation, under a low ATP concentration environment, CypA mainly promotes K48-linked pro-IL-1β ubiquitination at K132 and K143 sites to accelerate pro-IL-1β degradation.
Certainly, the researchers' study did not stop at this point, and follow-up utilizing a similar approach, through careful experimental design with comparison, the IL-1β-induced inflammation model was studied more systematically. It revealed that CypA exacerbated IL-1β-induced inflammatory mechanism, elucidated the role of CypA in IL-1β-induced type II epithelial-mesenchymal transition (EMT) for lung repair, providing a more systematic theoretical basis for future treatments of inflammation-related diseases, and the direction of relevant target screening.
In this research, we can see that a large number of knockout cell lines were used in almost all research methods. So the premade, good-quality knockout cell lines are useful tools to accelerate the research progress. Ubigene has established 3000+ KO cell lines which are now available, covering 8 major signaling pathways and 40 diseases, of which cell lines including THP-1, A549, 293T used in this paper (see table below). Ubigene can provide resources and services for scientific research. Promotional price starts from 1980 USD, as soon as 1-week delivery. Click here to search for in-stock KO cells >>











