[Research highlight] CRISPR-BTM knockout E.coli facilitates MMR sliding clamp
research

DNA mismatch repair (MMR) is a repair system after
genome replication, which plays an important role in improving DNA replication fidelity,
reducing gene mutation and maintaining genome stability. Mutations of MMR related genes in human
cells can inactivate the MMR system, increase the mutation rate of cells by more than 100 times,
and are associated with Lynch syndrome, hereditary nonpolyposis colorectal cancer (HNPCC) and
some sporadic cancers. Therefore, it is of great significance to further explore the molecular
mechanism of MMR system, which can provide a foundation for the prevention and treatment of
related diseases.
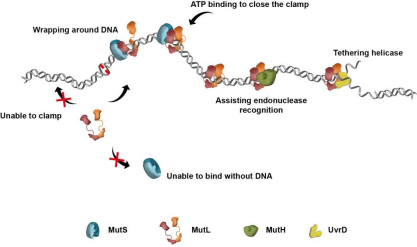
In E.coli's MMR system, after MutS recognizes the mismatch, it
will recruit MutL to form a complex, and then MutL will mediate the activation of downstream
proteins to repair the mismatch (Figure 1). From the recognition of mismatch to the
splicing of specific locus, the process of signal transmission needs to be efficient and
specific.
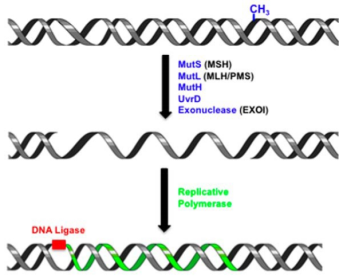
Figure 1. DNA mismatch repair process [1]
At
present, there are four models for mismatch repair initiation(Figure 2), which all believe that the mismatch signal is
transmitted by MutS-MutL complex. Besides the sliding model, the other three models all considered that the interaction between MutL and DNA has an
important function in the process of signal transmission.
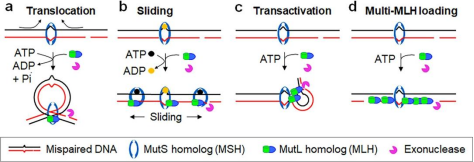
Figure 2. DNA mismatch repair initiation model [2]
However, the previous work of Jiaquan Liu’s team from the Center for Excellence in Molecular Cell
Science, CAS, found that MutL will undergo conformational changes
after ATP binding, forming MutL slicing clamps on DNA and spreading rapidly (Figure 3). The formation of
MutL sliding clamp is the premise of signal transmission. However, there
seems to be no continuous interaction between MutL sliding clamp and DNA.
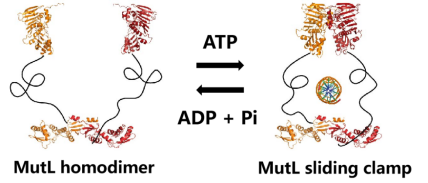
Figure 3. MutL structure
According to the literature, MutL interacts with DNA
through a positively charged cleft (PCC) in the N-terminal domain. Moreover, PCC mutation will
lead to a decrease of MMR system efficiency and an increase in cell mutation rate. In order to
explore the functions of key molecules and PCC in the process of MMR signal transmission, by
using single molecule fluorescence imaging technology and in vitro reconstruction of MMR system,
Jiaquan Liu's team, together with Richard Fishel's team from Ohio State University, published a
paper entitled "MutS functions as a clamp loader by positioning MutL on the DNA during mismatch
repair" in Nature Communications. This research result proves that MutS uses MutL PCC to promote
the formation of MutL sliding clamp on DNA, so as to transmit mismatch
signals.

Mutation of
PCC leads to the weakening of MutL's ability to bind DNA
By sequence comparison, the researchers found the
conserved amino acid sequences in PCC and mutated them. The result showed that PCC mutation
would weaken the ability of MutL to bind DNA (Figure 4).
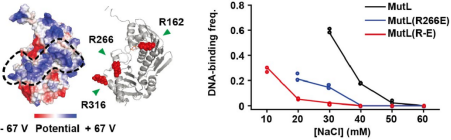
Figure 4. MutL and DNA interaction [3]
mutation result in inactivation of MMR system
In order to further explore whether the decreased
ability of MutL to bind DNA would lead to the inactivation of the MMR system, E.coli's MutL gene
knockout strain (provided by
Ubigene) was
used to carry out in vivo complementation experiment. They found that the mutation rate of
knockout strains could not be restored to the normal level by supplementing the mutated gene of
MutL PCC, which indicated that PCC mutation would lead to the inactivation of MMR system. At the
same time, the results of single molecule imaging show that the reason why PCC mutation causes
the deactivation of MMR system is that MutL mutant cannot form a complex with MutS (Figure
5).
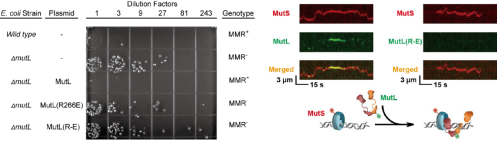
Figure 5. Inactivation of MMR system
caused by PCC mutation [3]
The CRISPR-B™ system
developed by Ubigene combines
the Red/ET recombination system with CRISPR/Cas9 gene editing system, and makes use of the advantages of
CRISPR/Cas9 system in efficient DNA cutting
and the further optimization of gene-editing
vector and gene-editing
process, which greatly improves the efficiency of bacterial gene editing, and can easily achieve the scarless
gene knockout and knockin
of strains. Escherichia
coli, Salmonella,
and Pseudomonas
aeruginosa are
all feasible. Positive
clones are delivered
as soon as 3 weeks. Click
to get
more details>>
MutS uses PCC to facilitate the loading of MutL sliding
clamp
To further study the effect of PCC mutation on the
assembly of MutS-MutL complex and the subsequent formation of MutL sliding clamp, the
researchers used the method of buffer exchange. First, partially recover the function of PCC
under the condition of low ionic concentration, and then replace the solution environment with
physiological ionic concentration to observe the formation of MutL sliding clamp. The results
showed that MutS activated and utilized MutL PCC, assembled with MutL and loaded MutL onto DNA
to form MutL sliding clamp.
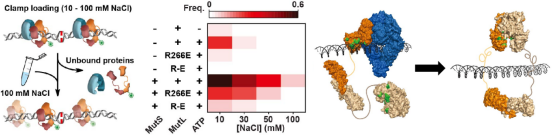
Figure 6. MutS utilizes MutL PCC to promote the loading of MutL sliding clamp [3]
MutS is a mismatch specific clamp loader
In the past, people believed that MutS would transmit
mismatch signals in the form of the complex after recruiting MutL. However, the researchers
found that the formation of MutL sliding clamp can be independent of MutS, and can activate
downstream pathways. Therefore, the function of MutS in MMR is only to load MutL onto DNA as a
mismatch specific clamp loader. Once MutL slipping clamp is formed, MutS is no longer required
to participate in the repair process downstream of MMR.
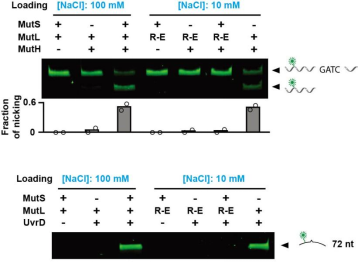
Figure 7. MutL sliding clamp activates MMR downstream
pathway [3]
This study proves that
MutL sliding clamp is the key molecule of MMR signal transmission, but
MutS is just a mismatch specific clamp loader, which also explains the function of MutL PCC, providing
new insight into the signal transmission mechanism of DNA mismatch repair.
Reference
[1] Fishel R. Mismatch repair. J Biol Chem. 2015 Oct
30;290(44):26395-403. doi: 10.1074/jbc.R115.660142. Epub 2015 Sep 9. PMID: 26354434; PMCID:
PMC4646297.
[2] Ortega, Janice et al. “Mispair-bound human
MutS-MutL complex triggers DNA incisions and activates mismatch repair.” Cell research vol. 31,5 (2021):
542-553. doi:10.1038/s41422-021-00468-y
[3] Yang, Xiao-Wen et al. “MutS functions as a clamp loader by
positioning MutL on the DNA during mismatch repair.”Nature communications vol. 13,1 5808. 3 Oct.
2022, doi:10.1038/s41467-022-33479-3















