knockout Liver Cancer Cell Line HepG2
Liver is vital to the body's metabolism, detoxification, blood coagulation and other essential processes. Participating in the body's immune system, the liver is an indispensable organ for maintaining the body's normal operation. In the study of liver diseases, one of the most useful tools is to create a suitable cell model. However, due to the difficulties in the obtainment and culturing for primary liver cells, the development of liver disease research has been confined in some ways. Moreover, the relatively high cost of culturing in primary liver cells also makes it less likely to carry out the research. Therefore, selecting liver cell lines with simple culture method and stable genetic background has become an alternative in liver research, and HepG2 is one of the most commonly used liver cancer cell lines.
The application of HepG2 cell line
HepG2 was established in 1979 by Knowles et al. It is a hepatoblastoma derived from a 15-year-old Caucasian male liver cancer specimen. HepG2 cells have an epithelial-like morphology, with a typical chromosome number of 55.
-
Hepatitis virus research: HepG2 does not contain hepatitis B virus (HBV) or hepatitis C virus (HCV), so HepG2 is a commonly used model for studies in HBV and HCV cell lines.
-
In vitro HCC model: HepG2 is a cell line derived from liver tissue of patients with hepatocellular carcinoma (HCC) and it is a common in vitro HCC model. There is no mutation in p53 tumor suppressor gene in HepG2, so it can be used to study the close relationship between p53 and hepatocellular carcinoma (HCC), as well as the onset, diagnosis, treatment and prevention of HCC.
-
Drug research: The liver is the most essential organ for detoxification and drug metabolism in the human body. HepG2 cell line is closer to human liver tissue both in terms of morphology and functions. Therefore, this cell line is often used in drug metabolism and liver toxicity studies.
The combination of liver cell line and CRISPR/Cas9 helps to study medical
problems such as hepatitis, genetic diseases and cancer
CRISPR/Cas9 mediated gene knockout HepG2 HBV infected model provides new ideas for cures of hepatitis B
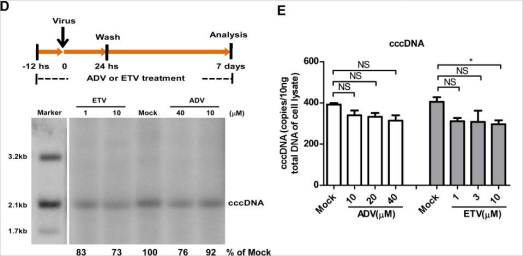
Hepatitis B virus (HBV) is a DNA virus. The genomic DNA of HBV outside the cell is double-stranded relaxed circular DNA (rcDNA). When the virus infects the cell, the HBV genome will enter the nucleus of the host cell. Then the rcDNA will be converted into covalently closed circular DNA (cccDNA) and exist stably in the cell. At present, an important reason for not being able to completely cure hepatitis B lies in the difficulties in clearing the stable cccDNA in the liver cell nucleus. The conversion of HBV rcDNA to cccDNA is a necessary step for the establishment of persistent infection of hepatitis B virus. Therefore, the study of the molecular mechanism of HBV cccDNA formation has far-reaching significance for the treatment of hepatitis B.
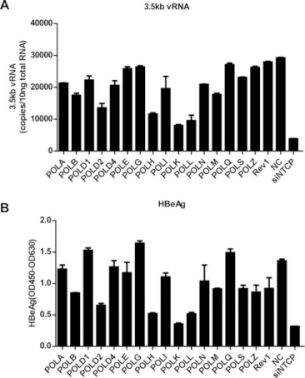
In order to explore the mechanism of HBV cccDNA formation, researchers used HepG2 to express the HBV receptor, the sodium ion-taurocholic acid co-transporter (NTCP). They established an efficient HepG2-NTCP cell model for HBV infection, which could detect the formation of cccDNA after only 24 hours. Afterwards, the experiments using viral DNA polymerase inhibitors and siRNA (targeting the host DNA polymerase) showed that HBV viral polymerase was not a critical factor in the conversion of rcDNA to cccDNA. However, the knockdown of POLK, POLL, and POLH can effectively reduce the expression levels of viral nucleocapsid protein (HBeAg) and 3.5 kb vRNA. Among three genes, the knockdown of POLK showed the most significant influence in the inhibition of HBV infection.
Furthermore, using CRISPR / Cas9 to knockout POLK in HepG2-NTCP cells can greatly decrease the formation of cccDNA, confirming that the formation of cccDNA in HBV requires the participation of POLK
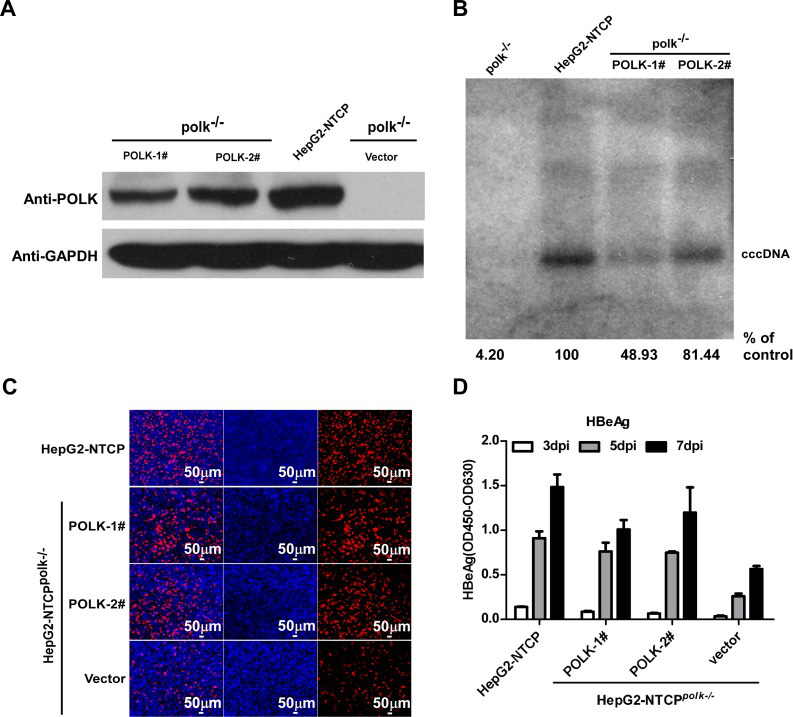
This study strongly indicated that POLK is a key cell molecule for HBV cccDNA formation in the HBV-infected HepG2-NTCP cell model, which provides new ideas for research in the molecular mechanism of cccDNA formation and new treatments for chronic hepatitis B.
CRISPR-U ™ can transfer CRISPR/Cas9 into Hep-G2 cells 10 times more efficiently, and select suitable cells for monoclonal culture after screening. Different clones were selected for target site amplification and sequencing verification, after which the positive clones with the knockout gene were selected.
Constructing HepG2 model of deletion mutation by CRISPR/Cas9 revealed the causes of Wilson disease
(hepatolenticular degeneration)
Hepatolenticular degeneration (Wilson disease) is a recessive genetic disorder caused by pathogenic loss-of-function variants in the ATP7B gene. It is characterized by disrupted copper homeostasis resulting in liver disease and/or neurological abnormalities. The variant NM_000053.3:c.1934T > G (Met645Arg) has been reported as compound heterozygous, and is highly prevalent among Wilson disease patients of Spanish descent. Accordingly, it is classified as pathogenic by leading molecular diagnostic centers. However, functional studies suggest that the amino acid change does not alter protein function, leading some researchers to question its pathogenicity. Here, researchers used a minigene system and gene-edited HepG2 cells to demonstrate that c.1934T > G causes ~70% skipping of exon 6.
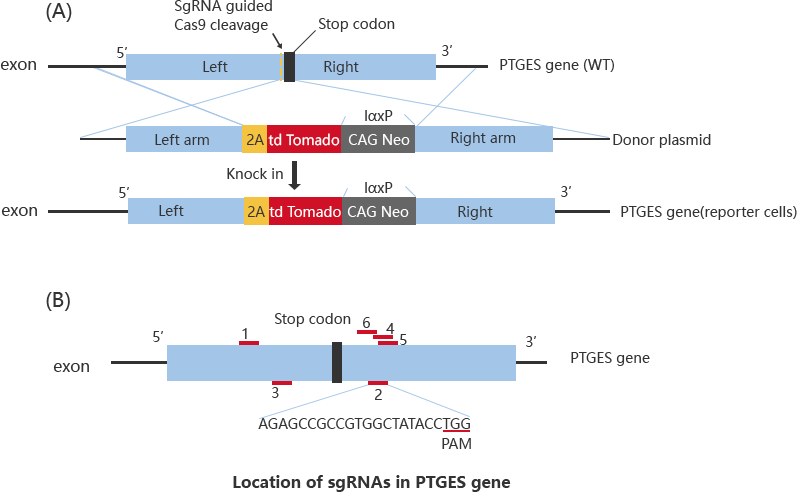
The CRISPR/Cas9 and ssODN were co-transfected into HepG2 cells by nuclear transfection and then selected for monoclonal culture after screening. The target sites were amplified and sequenced separately for the monoclonal, and the following 4 clones were screened for research.
|
Clone name
|
Genotype
|
|
2F3
|
Compound het c.1934T > G, large plasmid insertion
|
|
1F6
|
Homozygous c.1934T > G
|
|
1E8
|
Homozygous c.1934T > G
|
|
2A1
|
Homozygous c.1931dupA (chr13:52535987:C > CT)
|
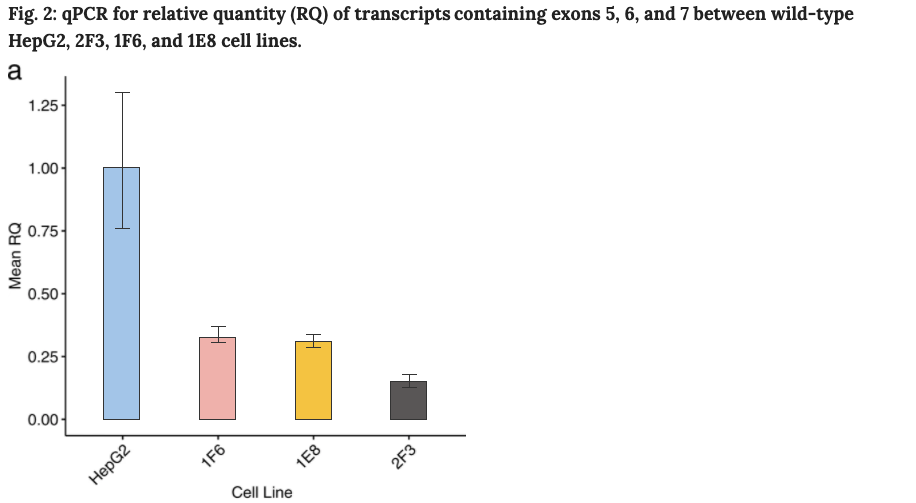
Compared to the transcription in wild-type cells, compound heterozygous HepG2 cells (2F3 clone) express only 15% of transcript containing exons 5, 6 and 7. Compared to wild-type cells, and homozygous cells (clones 1E8 and 1F6) express only 31–33%.

In addition, protein lysates were extracted from wild-type and edited HepG2 cells to determine ATP7B protein expression by western blot. Compared to wild-type cells, compound heterozygous (2F3) and homozygous (1E8, 1F6) cells express reduced levels of ATP7B, with increased ATP7B expression in 1E8 and 1F6 homozygous cell lines compared to 2F3 cells, as expected. A control 2A1 ATP7B knockout cell line has no detectable levels of ATP7B.
Moreover, the study suggested that c.1934T > G causes the skipping of exon 6, which results in frameshift and stop-gain, which leads to loss of ATP7B function. The outcome clarified the mechanism role of this point mutation in hepatolenticular degenerative diseases, which is helpful for the development of genetic drugs to restore correct splicing.
Fluorescent marker knockin HepG2 cells provide new technical support for anti-inflammatory and tumor treatment
mPGES-1 is a terminal rate-limiting enzyme responsible for inflammation-induced PGE2 production. The inhibition of mPGES-1 has been considered as a safe and effective target for the treatment of inflammation and cancer. However, a specific, efficient, and simple method for high-throughput screening of mPGES-1 inhibitors is still lacking. In this study, we developed a fluorescence imaging strategy to monitor the expression of mPGES-1 via CRISPR/Cas9 knock-in system.
By using nuclear transfection to efficiently transfer gRNA, Cas9 and Donor into HepG2 and other hepatocyte cell lines. After performing drug screening and monoclonal culture, the positive clones went through immunofluorescence co-localization, sequencing, RNAi and IL-1β treatment, which confirmed the successful construction of mPGES-1 reporter cells.
Fluorescent protein KI cells were treated with 4 common mPGES-1 inhibitors, after which the flow cytometry detected a significant attenuation of the fluorescent signal intensity. The result suggested that using the fluorescent protein KI cells is a high-efficiency and handy method in screening and optimization mPGES-1 inhibitor.
Meanwhile, the KI cells provide new technical support for anti-inflammatory and tumor treatments targeting small molecule compounds.
CRISPR-U™ efficiently modify genes in liver cell lines
CRISPR-U ™ is an exclusive technology independently developed by Ubigene Bioscience for gene editing cell lines. By optimizing gene editing vectors and processes, the efficiency in gene-cutting and recombination of CRISPR-U ™ is 10 times higher than the conventional CRISPR/Cas9 technology. We are capable of customizing genetically-modified liver cell lines as you desire and satisfying various needs in gene editing.
Reference:
Qi, Yonghe, et al. "DNA polymerase κ is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus." PLoS pathogens 12.10 (2016).
Merico, Daniele, et al. "ATP7B variant c. 1934T> G p. Met645Arg causes Wilson disease by promoting exon 6 skipping." NPJ Genomic Medicine 5.1 (2020): 1-7.
Chen, Zhanfei, et al. "CRISPR/Cas9-based liver-derived reporter cells for screening of mPGES-1 inhibitors." Journal of enzyme inhibition and medicinal chemistry 34.1 (2019): 799-807.