CRISPR Pseudomonas Aeruginosa,Gene Editing Service
Pseudomonas aeruginosa is a common encapsulated, Gram-negative, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes. This type of bacteria is found commonly in the environment, like in soil and in water. P. Aeruginosa is a gram-negative, aerobic rod bacterium of the Pseudomonadaceae family (a member of the Gammaproteobacteria). These years, P. aeruginosa are getting more popular in the field of CRISPR editing, which involves knockout cell lines, gene knock-in and point-mutation in the bacteria.
Applying CRISPR in P. aeruginosa Involving Insertion and Knockout of a Tag Gene
Pseudomonas aeruginosa is both a prototype multi-drug resistance (MDR) pathogen and a model species for CRISPR-Cas research. In a clinical setting that includes I-F CRISPR, this technology can be easily applied to the other two isolates of Pseudomonas aeruginosa. A two-step In-Del strategy was further developed, which involved inserting tags near the desired editing site, and then performing gene knockout to edit genomic sites lacking a valid PAM (protospacer adjacent motif) or essential genes. Among the three resistance mutations that enhance the resistance of fluoroquinolones, the gyrA mutation caused greater resistance compared with the efflux of MexAB-OprM or MexEF-OprN. These results promote the understanding of the development of MDR of clinical Pseudomonas aeruginosa strains and demonstrate the great potential of the natural CRISPR system in AMR research. Although there are complete genetic manipulation tools in various model strains, due to the diversity of DNA homeostasis and cytotoxicity of these strains, their application in "non-model" strains in medical, environmental and industrial senses is often affected. Hinder. Cloning of CRISPR-Cas9/Cpf1 system. The use of the natural CRISPR-Cas system with built-in genome targeting activity and widely distributed in prokaryotes provides a promising and effective method for solving these obstacles. The successful development of the first I-F CRISPR-mediated genome editing technology and its subsequent expansion to other clinical and environmental Pseudomonas aeruginosa isolates. Knockout Bacteria opens up a new approach to pathogen resistance functional genomics.
Silencing and Point Mutations in P. aeruginosa Helps Research in Bacterial Physiology, Drug Target Exploration, and Metabolic Engineering
Pseudomonas species exhibit significant biomedical, ecological, and industrial importance. Despite the extensive research and wide applications, genetic manipulation in Pseudomonas species, in particular in the major human pathogen Pseudomonas aeruginosa, remains a laborious endeavor. It is reported that a genome-editing method pCasPA/pACRISPR was developed by harnessing the CRISPR/Cas9 and the phage λ-Red recombination systems. The method allows for efficient and scarless genetic manipulation in P. aeruginosa. By engineering the fusion of the cytidine deaminase APOBEC1 and the Cas9 nickase, a base editing system pnCasPA-BEC was developed, which enables highly efficient gene inactivation and point mutations in a variety of Pseudomonas species, such as P. aeruginosa. Application of the two genome editing methods will dramatically accelerate a wide variety of investigations, such as bacterial physiology study, drug target exploration, and metabolic engineering.
The Rapid Construction of Gene Knockouts in P. aeruginosa with Base-pair Precision
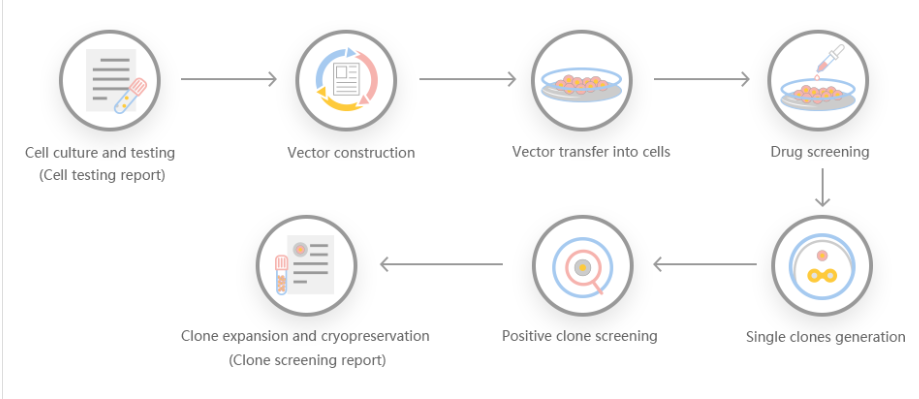
Pseudomonas aeruginosa is a model organism for the study of quorum sensing, biofilm formation, and also leading cause of nosocomial infections in immune-compromised patients. As such P. aeruginosa is one of the most well-studied organisms in terms of its genetics. However, the construction of gene KOs and replacements in Pseudomonas aeruginosa is relatively time-consuming, requiring multiple steps including suicide vector construction, conjugation, inactivation with the insertion of antibiotic resistance cassettes and allelic exchange. Even employing Gateway recombineering techniques with direct transformation requires a minimum of two weeks. Hence, a rapid streamlined method was developed to create clean KO mutants in P. aeruginosa through direct transformation, eliminating the need for the creation of Gateway-compatible suicide vectors. In this method, upstream and downstream sequences of the gene/locus to be deleted are amplified by polymerase chain reaction (PCR) and seamlessly fused with the linearized pEX18Tc sacB suicide plasmid by Gibson assembly. The resulting knockout plasmid is transformed into P. aeruginosa by an electroporation method optimized in this study. The plasmid is then integrated into the chromosome by homologous recombination, and knockout mutants are identified via sacB mediated sucrose counter-selection. The current method was employed to generate clean gene knockouts of the heme assimilation system anti-σ factor, hasS, and the virulence regulator involving ECF system anti-σ and σ factors vreA and vreI, respectively. The process from plasmid construction to confirmation by DNA sequencing of the gene knockout was completed in one week. Furthermore, the utility of the method is highlighted in the construction of the vreA and vreI knockouts, where the start codon of vreA and the stop codon of vreI overlap. Utilizing Gibson assembly knockout mutants were constructed with single base-pair precision to generate the respective vreA and vreI knockouts while maintaining the start and stop codon of the respective genes. Overall, this method allows for the rapid construction of gene KOs in P. aeruginosa with base-pair precision.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
References:
Zeling Xu, Ming Li, Yanran Li, Huiluo Cao, Hua Xiang, Aixin Yan. Native CRISPR-Cas mediated in situ genome editing reveals extensive resistance synergy in the clinical multidrug resistant Pseudomonas aeruginosa. bioRxiv 496711.
Weizhong Chen, Ya Zhang, Yifei Zhang, Yishuang Pi, Tongnian Gu, Liqiang Song, Yu Wang, Quan jiangJi. iScience, Volume 6, 31 August 2018, Pages 222-231.
Huang, Weiliang, and Angela Wilks. “A rapid seamless method for gene KO in Pseudomonas aeruginosa.” BMC microbiology vol. 17,1 199. 19 Sep. 2017, doi:10.1186/s12866-017-1112-5.
Ubigene Biosciences is co-founded by biological academics and elites from China, the United States, and France. We are located in Guangzhou Science City, which serves as a global center for high technology and innovation. Ubigene Biosciences has 1000㎡ office areas and laboratories, involving genome editing, cell biology technology, and zebrafish research. We provide products and services for plasmids, viruses, cells, and zebrafish. We aim to provide customers with better gene-editing tools for cell or animal research.
We developed CRISPR-U™ and CRISPR-B™(based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, CRISPR-U™ and CRISPR-B™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo.
Genome Editing Platform
——Focusing on the Application of CRISPR-U™ and CRISPR-B™ Gene Editing Technology
1. Provides various types of gene-editing vectors for different species.
2. Provides different virus packaging services, including lentiviruses, adenoviruses and adeno-associated viruses.3. Provides high-quality services for gene knockout, point mutation and knockin cell lines.
Cell Biology Platform
——Focusing on primary cell
1. Provides over 400 types of primary cells.
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.






