Location: Home > Gene Editing Services > Microbe > Gene-editing Microorganisms
Traditional bacterial gene editing mainly uses the RecBCD system that widely exists in bacteria to recombine the suicide plasmid with the genome to knock out the target gene. However, the efficiency of this method is extremely low, which requires repeated experiments, and may also lead to reverse mutation, and it is difficult to obtain the target gene knockout strains. λ-Red is a better choice for Escherichia coli and its close species. Its recombination efficiency will be slightly higher than the endogenous recombination system in bacteria, but the target gene needs to be replaced with a resistance gene. After successful replacement, although the recombinant enzyme can be transferred to bacteria to delete the resistance gene, it will still leave a recombination site on the genome.
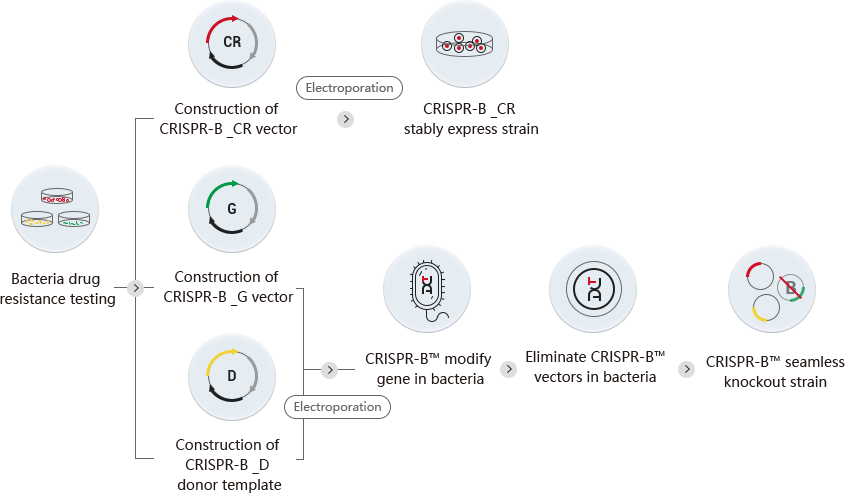
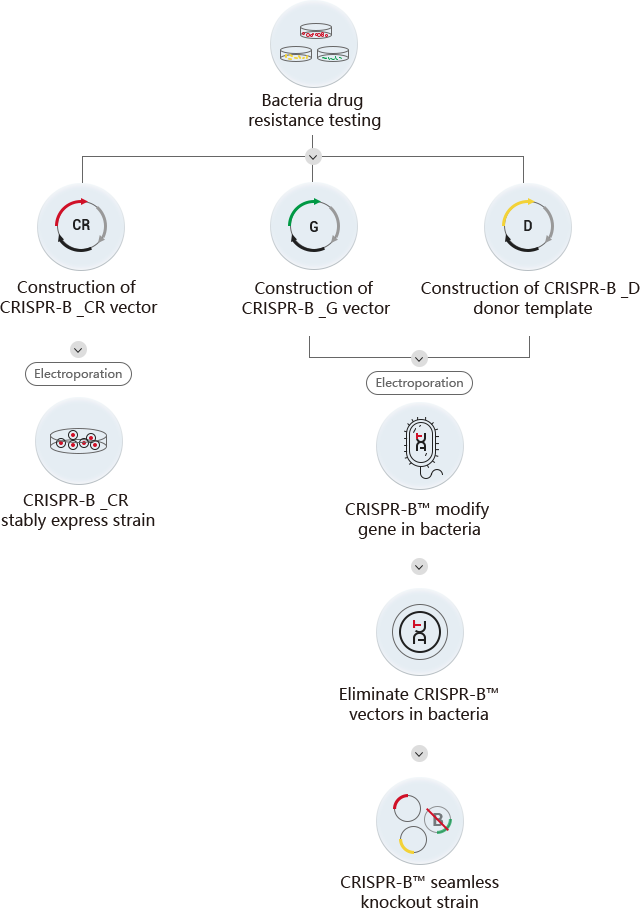
Based on the above problems, the CRISPR-B™ technology system developed by Ubigene innovatively combines the Red/ET recombination system with CRISPR/Cas9 gene editing system, uses the advantages of CRISPR/Cas9 system in efficient cutting and further optimizes the gene editing plasmid and gene editing process, greatly improving the efficiency of bacterial gene editing. Compared with the previous two methods, the CRISPR-B™ technology of Ubigene can not only achieve high efficiency, but also achieve scarless knockout (that is, no residual resistant gene or recombination sites, and eliminate foreign plasmids at the same time). In addition, this technology can achieve target gene point mutation and knockin which cannot be achieved by the previous two methods. At present, Ubigene can provide gene editing (knockout/point mutation/knockin) and construction of overexpression strains of Escherichia coli, Salmonella and Pseudomonas aeruginosa, guaranteeing the delivery of positive homozygous clones.
The efficiency is 20-30 times higher than that of the classic methods;
Scarless gene-editing technology, safe and sound;
Easily achieve microbial gene knockout (KO), point mutation (PM) and knockin (KI);
It is possible to knockout multiple genes simultaneously.
| CRISPR-B™ vector construction | |
| CRISPR-B _CR | Carrying Cas9 nuclease and Red recombinase |
| CRISPR-B _G | Carrying target gene gRNA |
| CRISPR-B _D | Carrying donor template |