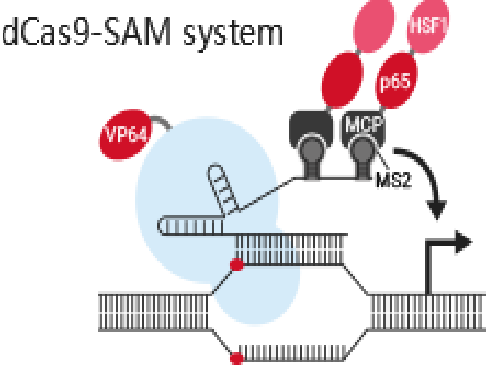
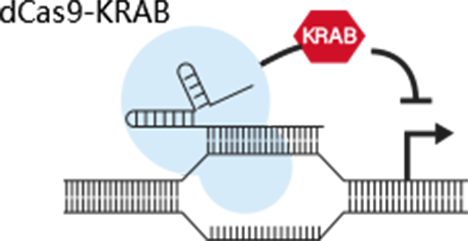
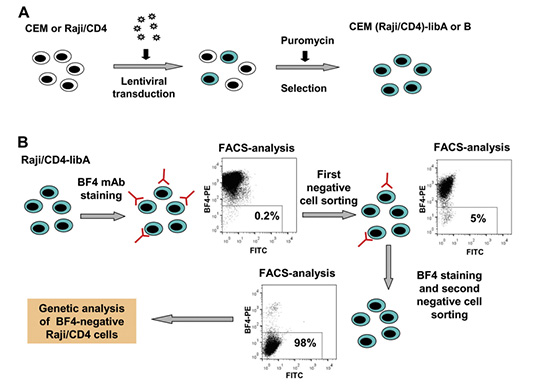
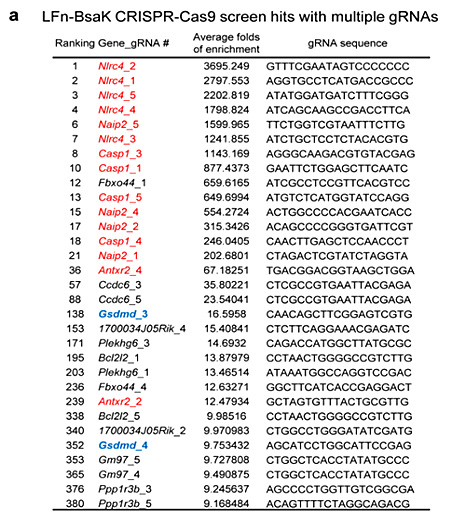
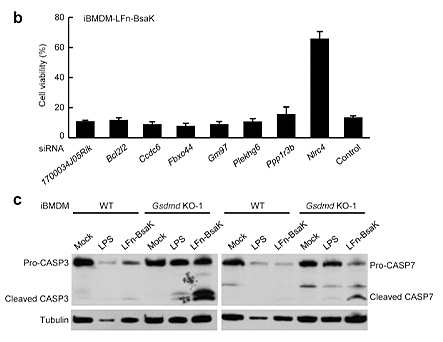
References
[1]Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE,
Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014
Jan 3;343(6166):84-87. doi: 10.1126/science.1247005. Epub 2013 Dec 12. PMID: 24336571; PMCID:
PMC4089965.
[2]Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon
K, Klepper K,
Pekin D, Beckett AN, Sanchez KS, Thomas A, Kuo CC, Du D, Roguev A, Lewis NE, Chang AN, Kreisberg
JF, Krogan N, Qi L, Ideker T, Mali P. Combinatorial CRISPR-Cas9 screens for de novo mapping of
genetic interactions. Nat Methods. 2017 Jun;14(6):573-576. doi: 10.1038/nmeth.4225. Epub 2017
Mar 20. PMID: 28319113; PMCID: PMC5449203.
[3]Park RJ, Wang T, Koundakjian D, Hultquist JF,
Lamothe-Molina P, Monel B, Schumann K, Yu H,
Krupzcak KM, Garcia-Beltran W, Piechocka-Trocha A, Krogan NJ, Marson A, Sabatini DM, Lander ES,
Hacohen N, Walker BD. A genome-wide CRISPR screen identifies a restricted set of HIV host
dependency factors. Nat Genet. 2017 Feb;49(2):193-203. doi: 10.1038/ng.3741. Epub 2016 Dec 19.
PMID: 27992415; PMCID: PMC5511375.
[4]Zotova A, Zotov I, Filatov A, Mazurov D. Determining
antigen specificity of a monoclonal
antibody using genome-scale CRISPR-Cas9 knockout library. J Immunol Methods. 2016 Dec;439:8-14.
doi: 10.1016/j.jim.2016.09.006. Epub 2016 Sep 21. PMID: 27664857.
[5]Shi J, Zhao Y, Wang K,
Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of
GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015 Oct
29;526(7575):660-5. doi: 10.1038/nature15514. Epub 2015 Sep 16. PMID: 26375003.