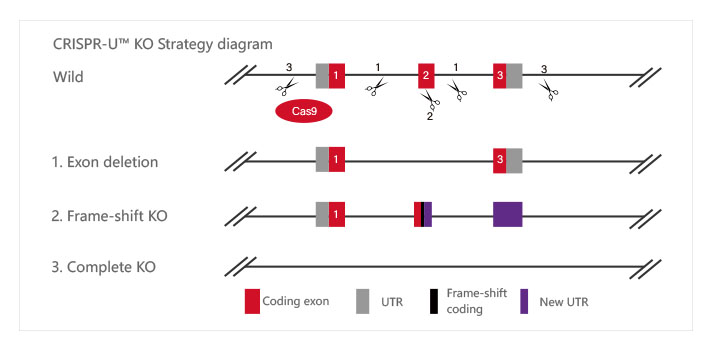
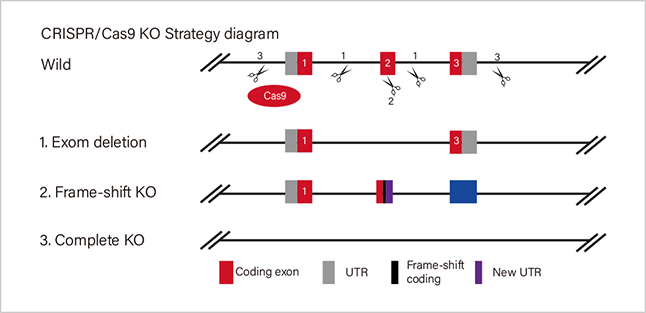
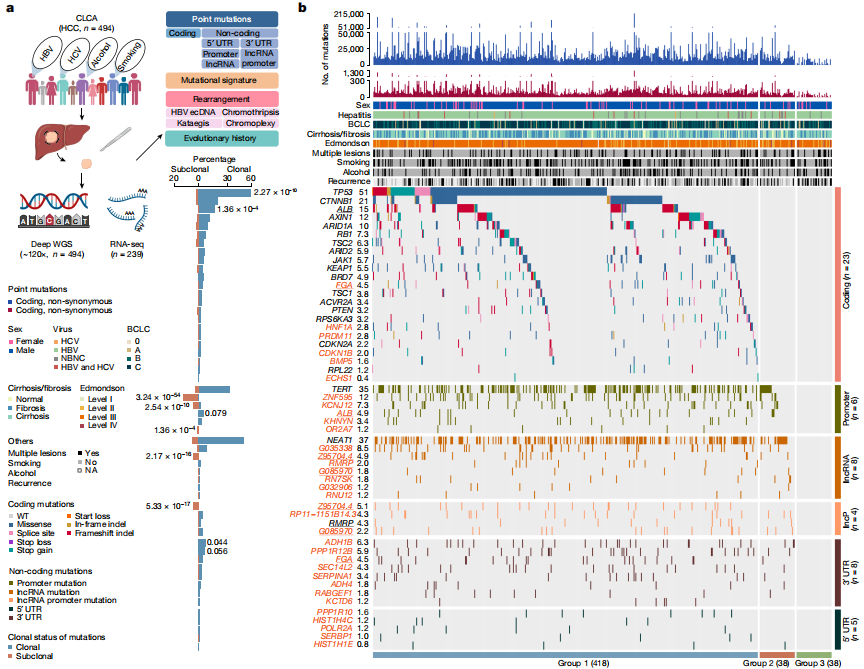
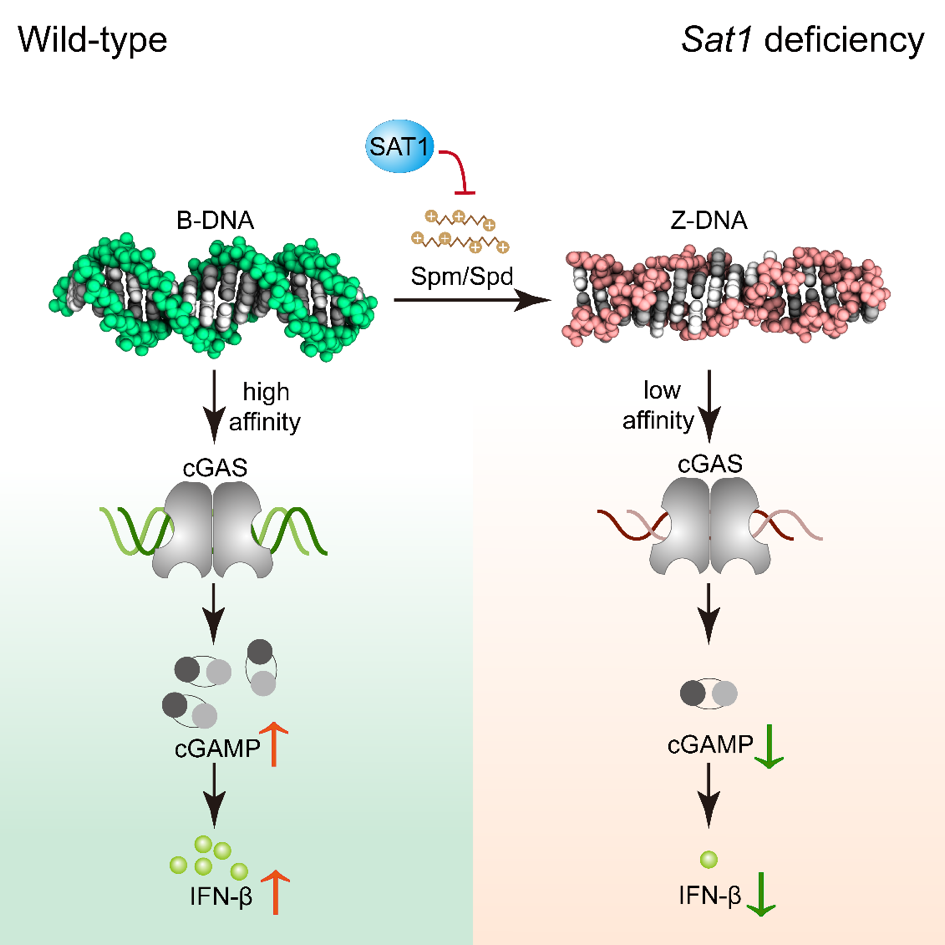
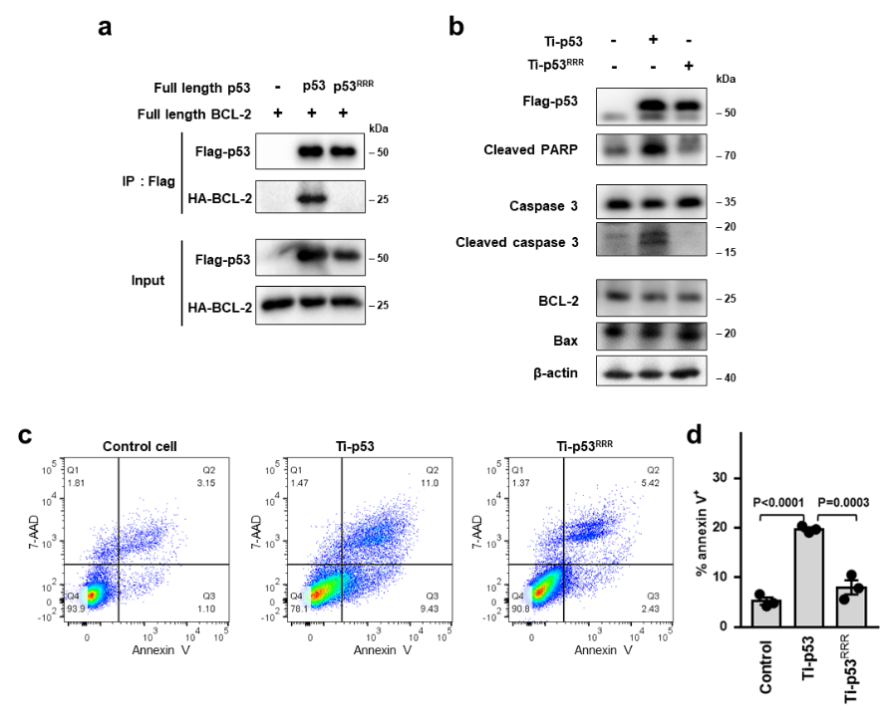
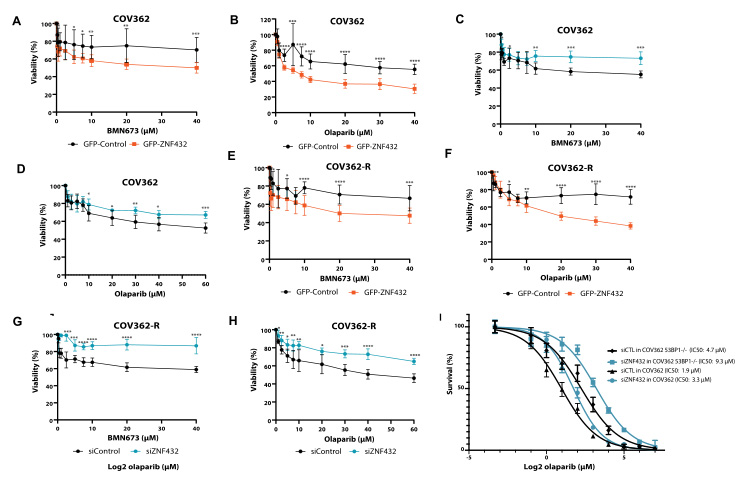
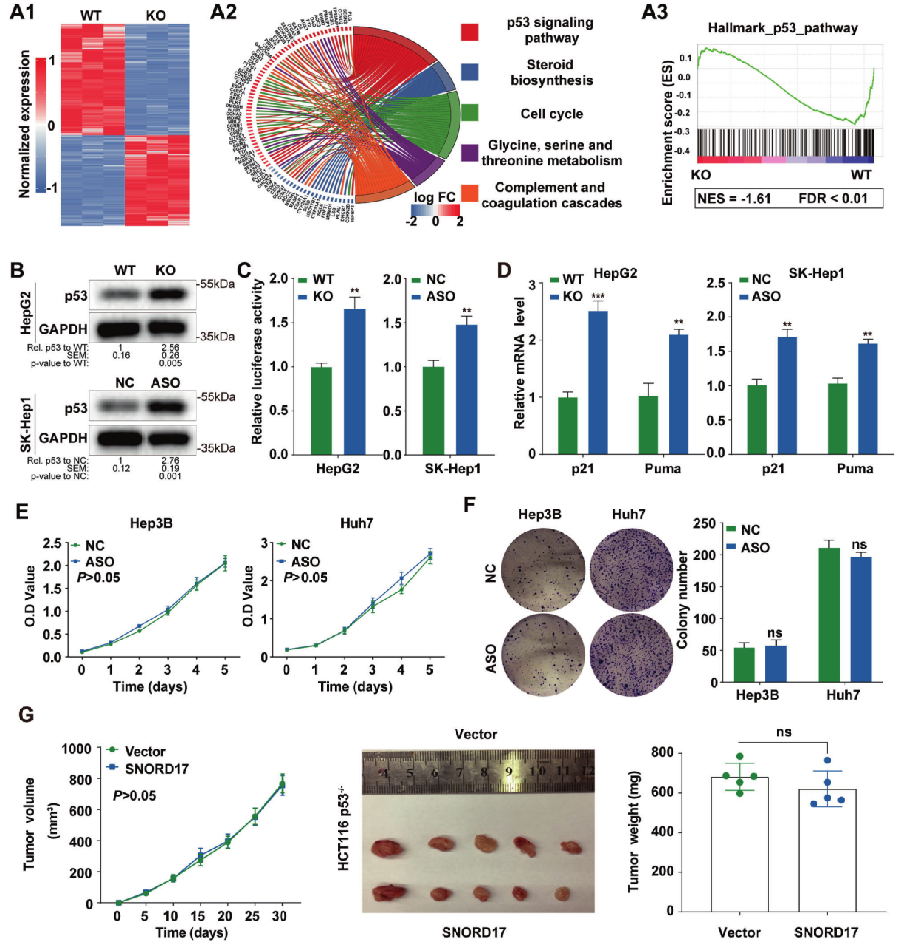
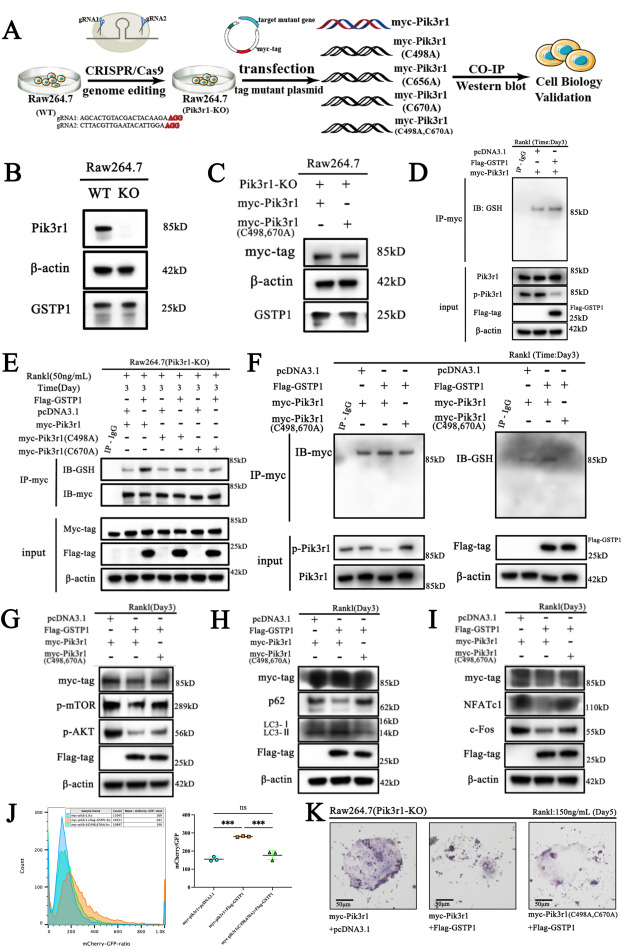
ZNF protein was confirmed as a key factor regulating the genome integrity of
mammalian cells. In order to explore the possibility that ZFP can be used as an
effector of DNA repair based on homologous recombination (HR), Jean Yves Masson
[3] team of Laval University used the
ZNF432 knockout U2OS cell line constructed by Ubigene to carry out a series of experiments, and found that ZNF432 deletion in cancer cells
would accelerate DNA repair, lead to the weakening of PARPi effect, and make ovarian
cancer cells develop drug resistance, confirming that ZNF432 is a new HR inhibitor,
which successfully broadened the new way to study the efficacy of PARPi.
View details>> 



 Circulatory System
Circulatory System Endocrine System
Endocrine System Brain and Nervous System
Brain and Nervous System Blood and lymphatic System
Blood and lymphatic System Urinary System
Urinary System Digestive System
Digestive System

 Reproductive System
Reproductive System Stem Cell
Stem Cell