
A human leukemic cell line (THP-1) cultured from the blood of a boy with acute monocytic leukemia. Since its establishment in 1980, THP-1 cells have been widely used in the research of monocyte and macrophage related mechanisms, signaling pathways, nutrient and drug transportation. The morphological and functional characteristics of THP-1 are very similar to human primary monocytes (including cell differentiation markers). Compared with human peripheral blood monocytes (PBMC), THP-1 is easier to be cultured and has a more consistent background. Therefore, THP-1 is a commonly used acute monocytic leukemia cell line in various laboratories, and it is an ideal tool for studying immunity and inflammation.
Application of THP-1: macrophage and inflammation model
THP-1can be differentiated into M1/M2 macrophages and release corresponding cytokines.
M1 macrophage polarization:
-
THP-1 can be induced to differentiate into macrophages by Phorbol 12-myristate 13-acetate (PMA), and then M1 polarization can be induced by lipopolysaccharide (LPS) and IFN -γ, releasing TNF -α, IL-6 and other cytokines.This is a typical inflammatory model.
M2 macrophage polarization:
-
M2 polarization can beinduced by IL-4, IL-13 and macrophage colony-stimulating factor (M-CSF). TGF - β, IL-10 and other inhibitory cytokines can be released. This is similar to the process of tissue repair and reconstruction in the late stage of inflammation.
Atherosclerotic inflammation model:Under the action of oxidized low-density lipoprotein (ox-LDL), macrophages can further become foam cells. This is a pathological cell in atherosclerotic plaques and is a chronic inflammation model.
The combination of THP-1 and CRISPR/Cas9 technology is helpful for the study of immune and inflammatory diseases
THP-1 is a near-tetraploid suspension cell, and the success rate of THP-1 is very low by conventional gene-editing methods. CRISPR/Cas9 is widely used to construct gene-editing THP-1 model because of its simple, high efficiency and low toxicity.
Key genes for macrophagesclearing pathogens was found by CRISPR/Cas9 mediated gene knock-out THP-1 model
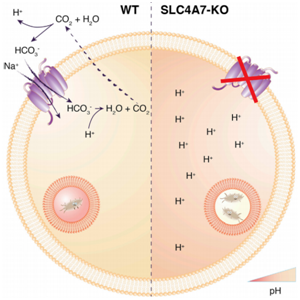
Phagosome acidification of macrophages is an essential step to eliminate pathogens. Phagosome acidification is closely related to the metabolism of macrophages and the transportation of nutrients. And the transportation of metabolites is closely related to solute carrier (SLC)protein.The researchers found that the bicarbonate transporter SLC4A7 in the SLC family is an essential gene for phagosome acidification of macrophages. In CRISPR/Cas9 mediated SLC4A7 knockout THP-1 cell line, the ability of phagosome acidification and killing bacteria was reduced. The acidity of the phagocyte was increased after the supplementation of SLC4A7. This indicates that SLC4A7 mediated bicarbonate driven in macrophages is essential for the maintenance of cytoplasmic pH and phagosome acidification. CRISPR-UTM can efficiently transfer gRNA and Cas9 into THP-1 cells by nucleofection.After drug screening, single clones would be generated. Positive clones would be validated by sequencing.
CRISPR/Cas9 mediated Chronic granulomatous disease (CGD) THP-1 cell line model is helpful to develop better disease treatments
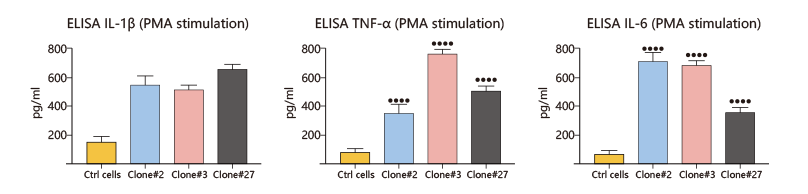
Chronic granulomatous disease (CGD) is a rare X-linked genetic disease. Due to the mutation or deficient of CYBB gene, macrophages lack nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, and cannot produce hydrogen peroxide to effectively kill the invading microorganisms. This usually leads to serious repeated infections caused by bacteria, fungi and other microorganisms. Some researchers used CRISPR/Cas9 technology to knockout CYBB gene and generate point mutation c.90c>G (found in a CGD patient) in THP-1 cells, and successfully constructed the CGD model. Compared with wild-type THP-1 cells, two KO clones (#3 and #27) and a point mutation clone (#2, c.90c > G) showed decrease in H2O2 level after PMA and LPS induction, and a significant increase in IL-1β, TNF-α and IL-6 release, which was consistent with the behavior of macrophages in CGD. This CGD model provides a powerful tool for disease study and will help to develop better treatments.
By using CRISPR-UTM,THP-1 cell line would be efficiently co-transfected with gRNA, Cas9 and ssODN. After drug screening, single clones would be generated. Positive clones would be validated by sequencing.
The signal pathway of intracellular antiviral response was confirmed by the THP-1 Cell Models of gene knock-out and knock-in.
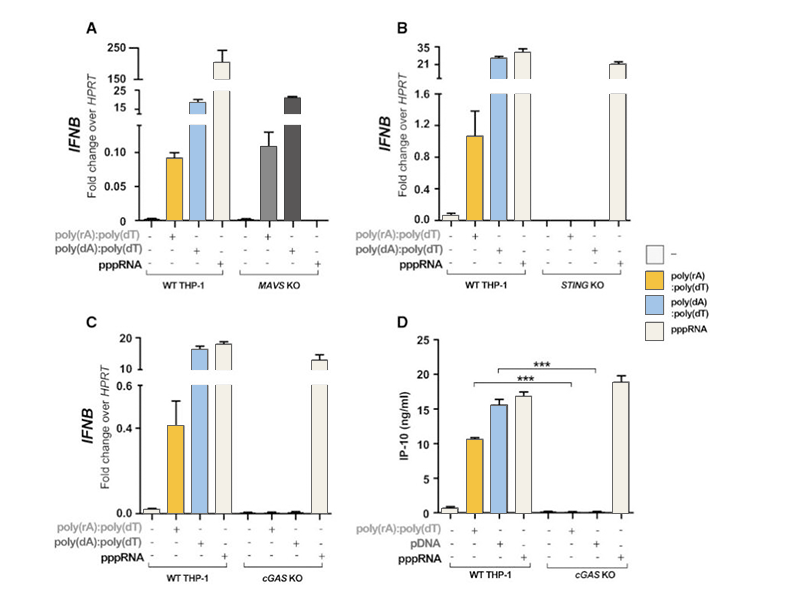
The abnormal location of DNA in the cytoplasm is usually related to virus infection or tumor. The cGAS-cGAMP-STING pathway can detect the existence of cytosolic dsDNA, and induce a strong immune response, producing interferon and activating other immune response genes. RIG1-MAVS can detect pppRNA (dsRNA, the genome of some viruses) in cytoplasm and induce immune response. Sometimes there is a complex of RNA and DNA in the cytoplasm, which usually occurs in the case of some virus infection. In order to study which pathway that the RNA-DNA complex activates the immune response, the researchers generatedMAVS, cGAS,STING knockout THP-1 cell lines, and introduced dsDNA, ppRNA and RNA-DNA complex into the cells respectively. It was found that the RNA-DNA complex is activated by the cGAS-cGAMP-STING pathway.
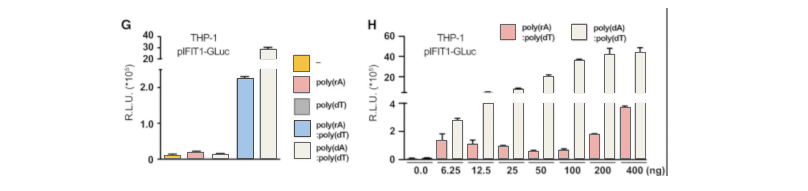
Then, the researchers used CRISPR/Cas9 technology to insert2A-GLuc into the IFIT1 gene. IFIT1 is a typical interferon activated gene. Subsequent experiments showed that after the introduction of RNA-DNA complex, the expression of Gluc was driven by the activation of IFIT1 promoter due to the expression of interferon. These results further proved that the RNA-DNA complex in the cytoplasm activated the immune response through the cGAS-cGAMP-STING pathway.By using CRISPR-UTM,THP-1 cell line would be co-transfected with gRNA, Cas9 and donor vector. After drug screening, single clones would be generated. Positive clones would be validated by sequencing.
CRISPR-U™ efficiently modify genes in THP-1 cell line. CRISPR-U™, developed by Ubigene, is more efficient than general CRISPR/Cas9 in double-strand breaking, and CRISPR-U™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI). Ubigene can customize the gene-editing THP-1 cell line and other monocytes that you are interested in. We can also help you to generate various genes overexpression in THP-1 cell line.
References:
[1]Tsuchiya S, Yamabe M, Yamaguchi Y, et al. Establishment and characterization of a human acute monocytic leukemia cell line (THP‐1)[J]. International journal of cancer, 1980, 26(2): 171-176.
[2] Chanput W, Mes J J, Wichers H J. THP-1 cell line: an in vitro cell model for immune modulation approach[J]. International immunopharmacology, 2014, 23(1): 37-45.
[3] Sedlyarov V, Eichner R, Girardi E, et al. The bicarbonate transporter SLC4A7 plays a key role in macrophage phagosome acidification[J]. Cell host & microbe, 2018, 23(6): 766-774. e5.
[4] Benyoucef A, Marchitto L, Touzot F. CRISPR gene-engineered CYBBko THP-1 cell lines highlight the crucial role of NADPH-induced reactive oxygen species for regulating inflammasome activation[J]. Journal of Allergy and Clinical Immunology, 2020.
[5] Mankan A K, Schmidt T, Chauhan D, et al. Cytosolic RNA: DNA hybrids activate the cGAS–STING axis[J]. The EMBO journal, 2014, 33(24): 2937-2946.










