The Arsenal for Finding Ferroptosis Targets - CRISPR Library

Ferroptosis, a novel form of cell death first proposed in 2012[1], has attracted widespread attention since its discovery, appearing in various research fields such as neurodegenerative diseases, stroke, and cancer. Exploring the regulatory mechanisms of ferroptosis to find new therapeutic targets for diseases has become one of the current research hotspots. CRISPR library, as a powerful tool for target screening, has long been shining in the field of ferroptosis research. Today, let’s walk through how CRISPR library is applied in the research of ferroptosis target screening.
Mechanism of Ferroptosis Occurrence
How ferroptosis occurs? As the name suggests, ferroptosis is an iron ion-dependent cell death pathway. When extracellular trivalent iron ions Fe3+ are transported into the cytoplasm through transferrin, Fe3+ is reduced to ferrous ions Fe2+; Under normal circumstances, Fe2+ will preferentially form Fe2+ complexes, and excess Fe2+ will be transported to the extracellular space through ferrous transporters. When the Fe2+ content is overloaded, excess Fe2+ cannot be discharged from the cell in a timely manner and accumulates in the cytoplasm. Free Fe2+ further generates a large amount of reactive oxygen species (ROS) through the Fenton reaction, which can cause unsaturated fatty acid peroxidation on the cell membrane, leading to loss of cell membrane function and membrane rupture, ultimately leading to cell death[2-4].
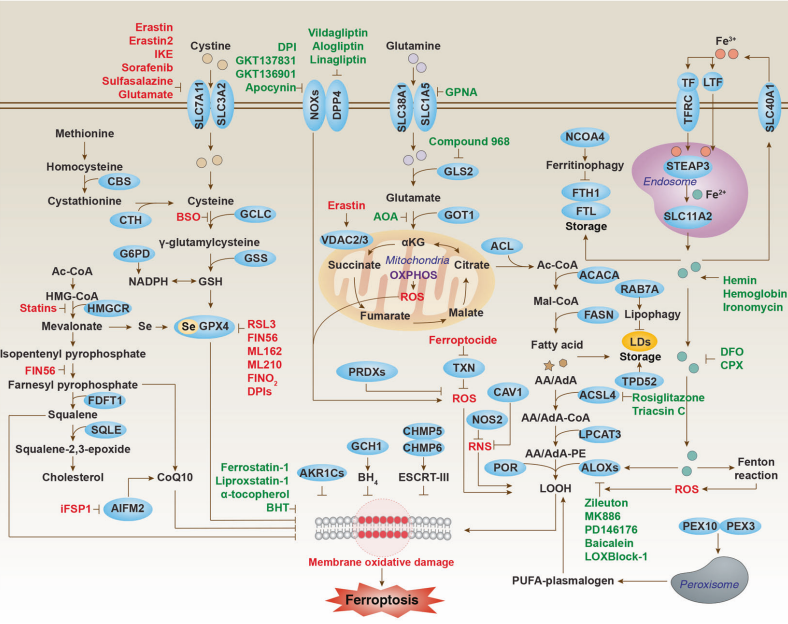
Figure 1. Ferroptosis-regulated signaling pathway[4]
Essentially, the occurrence of ferroptosis is due to the imbalance of the intracellular redox system and the high content of peroxides, leading to the oxidation of unsaturated fatty acids on the cell membrane, causing damage to the cell membrane function and even rupture. Glutathione peroxidase 4 (GPX4), with its efficient membrane lipid peroxide clearance capability, was the first identified ferroptosis inhibitor and is considered the core regulatory factor of ferroptosis. With the development of ferroptosis research, especially the emergence of CRISPR library screening technology, more and more ferroptosis regulatory factors have been discovered. Genes such as FSP1, GCH1, and the latest reported DHCR7 have all been discovered through CRISPR library screening. Let's introduce them one by one.
CRISPR Synthetic Lethal Library Screening Reveals FSP1
In some GPX4 knockout tumor cell lines, there are still
cells resistant to ferroptosis, indicating that besides GPX4, there are other pathways regulating ferroptosis. To identify other ferroptosis resistance genes, Bersuker et al. conducted synthetic lethal library screening on osteosarcoma
cells U2 OS. After treating the U2 OS library cells with the GPX4 inhibitor RSL3, negative screening revealed a
significant loss of FSP1, indicating that the knockout of the FSP1 gene is highly sensitive to ferroptosis, meaning that the expression of FSP1 inhibits cell ferroptosis. Further
experiments by researchers have found that FSP1 promotes the resistance of cancer cells to ferroptosis by producing antioxidant forms of CoQ10[5].
With the deepening of research on ferroptosis, more and more articles related to FSP1 and ferroptosis have been
published, which also proves the reliability of CRISPR library screening targets.
![CRISPR synthetic lethal library screening revealed FSP1[5] CRISPR synthetic lethal library screening revealed FSP1[5]](/uploads/allimg/240318/1-24031Q5320O50.png)
Figure 2. CRISPR synthetic lethal library screening revealed FSP1[5]
GCH1/BH4 Signaling Pathway Inhibits Ferroptosis
In addition to the FSP1/CoQ10 signaling pathway mentioned above that inhibits ferroptosis, GCH1/BH4 is also one of the signaling pathways that inhibit ferroptosis discovered through CRISPR library screening in recent years. Kraft et al. treated mouse fibroblasts with three different ferroptosis induction methods (RSL3, IKE, and GPX4 gene knockout) and conducted genome-wide activation library screening. By comparing the enrichment information of sgRNA in the three different treatment methods, the researchers found that only the GCH1 gene was significantly enriched under these three treatment conditions. Further research on the mechanism of GCH1 inhibiting ferroptosis revealed that GCH1 suppresses cell ferroptosis by promoting the expression of the antioxidant BH4[6]. Similarly, Soula et al. used a metabolic gene knockout library to screen for ferroptosis resistance genes in Jurkat cells, and also found significant enrichment of the GCH1 gene and other genes related to BH4 synthesis, verifying the inhibitory effect of the GCH1/BH4 signaling pathway on ferroptosis in Jurkat cells[7].
![Both the screening using the genome-wide activation library (A)[6] and metabolic gene knockout library (B)[7] found the enriched GCH1 Both the screening using the genome-wide activation library (A)[6] and metabolic gene knockout library (B)[7] found the enriched GCH1](/uploads/allimg/240318/1-24031Q53441Q2.png)
Figure 3. Both the screening using the genome-wide activation library (A)[6] and metabolic gene knockout library (B)[7] found the enriched GCH1
Regulation of Ferroptosis by 7-DHC ContentRecently, a team led by Professor Wang Ping from Tongji University in China published an article in Nature describing the screening of ferroptosis targets through genome-wide knockout libraries. The team treated 293T library cells with RSL3 and found through sequencing analysis that genes in the distal cholesterol synthesis pathway were significantly enriched. Among them, the genes MSMO1, CYP51A1, EBP, and SC5D have anti-ferroptosis effects, while DHCR7 promotes ferroptosis. In the distal cholesterol synthesis pathway, the genes MSMO1, CYP51A1, EBP, and SC5D promote the synthesis of 7-dehydrocholesterol (7-DHC), while DHCR7 promotes the conversion of 7-DHC to cholesterol, indicating that the 7-DHC content may be a key regulator of ferroptosis. Further research on the relationship between 7-DHC and ferroptosis revealed that 7-DHC acts as an antioxidant in ferroptosis, inhibiting phospholipid peroxidation at the cost of its own oxidation, thereby preventing ferroptosis[8]. It is worth mentioning that an accompanying article published by Freitas et al. in Nature also pointed out that 7-DHC is an endogenous ferroptosis inhibitor, confirming the accuracy of the library screening results[9].
![The genome-wide knockout library revealed the relationship between distal cholesterol synthesis pathway and ferroptosis[8] The genome-wide knockout library revealed the relationship between distal cholesterol synthesis pathway and ferroptosis[8]](/uploads/allimg/240318/1-24031Q53GBZ.png)
Figure 4. The genome-wide knockout library revealed the relationship between distal cholesterol synthesis pathway and ferroptosis[8]
In fact, the ferroptosis targets
identified through CRISPR library screening are far more than those mentioned above. For example, the team led by
Xuejun Jiang discovered through genome-wide activation library screening
that the MBOAT2 gene regulates the composition of unsaturated fatty acids in cells, inhibiting ferroptosis[10].
Ryan et al. used a genome-wide knockout library to screen neural cells and
found that the vesicle transport gene SEC24B promotes ferroptosis in
oligodendrocytes[11], and so on.
The genes discovered so far are just the tip of the iceberg in the ferroptosis regulatory network, and different cells may have their
unique ferroptosis regulatory mechanisms. From the above cases, it is
easy to see that using CRISPR library screening for targets has high precision, good repeatability, and the
combination of CRISPR library with ferroptosis has produced many
high-scoring articles. If you have a need to find ferroptosis targets,
hurry up and use the CRISPR library!
References
[1] Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012 May 25;149(5):1060-72.
[2] Bai Q, Liu J, Wang G. Ferroptosis, a Regulated Neuronal Cell Death Type After Intracerebral Hemorrhage. Front Cell Neurosci. 2020 Nov 16;14:591874.
[3] Stockwell BR. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022 Jul 7;185(14):2401-2421.
[4] Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021 Feb;31(2):107-125.
[5] Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, Bassik MC, Nomura DK, Dixon SJ, Olzmann JA. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019 Nov;575(7784):688-692.
[6] Kraft VAN, Bezjian CT, Pfeiffer S, Ringelstetter L, Müller C, Zandkarimi F, Merl-Pham J, Bao X, Anastasov N, Kössl J, Brandner S, Daniels JD, Schmitt-Kopplin P, Hauck SM, Stockwell BR, Hadian K, Schick JA. GTP Cyclohydrolase 1/Tetrahydrobiopterin Counteract Ferroptosis through Lipid Remodeling. ACS Cent Sci. 2020 Jan 22;6(1):41-53.
[7] Soula M, Weber RA, Zilka O, Alwaseem H, La K, Yen F, Molina H, Garcia-Bermudez J, Pratt DA, Birsoy K. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat Chem Biol. 2020 Dec;16(12):1351-1360.
[8] Li Y, Ran Q, Duan Q, Jin J, Wang Y, Yu L, Wang C, Zhu Z, Chen X, Weng L, Li Z, Wang J, Wu Q, Wang H, Tian H, Song S, Shan Z, Zhai Q, Qin H, Chen S, Fang L, Yin H, Zhou H, Jiang X, Wang P. 7-Dehydrocholesterol dictates ferroptosis sensitivity. Nature. 2024 Feb;626(7998):411-418.
[9] Freitas FP, Alborzinia H, Dos Santos AF, Nepachalovich P, Pedrera L, Zilka O, Inague A, Klein C, Aroua N, Kaushal K, Kast B, Lorenz SM, Kunz V, Nehring H, Xavier da Silva TN, Chen Z, Atici S, Doll SG, Schaefer EL, Ekpo I, Schmitz W, Horling A, Imming P, Miyamoto S, Wehman AM, Genaro-Mattos TC, Mirnics K, Kumar L, Klein-Seetharaman J, Meierjohann S, Weigand I, Kroiss M, Bornkamm GW, Gomes F, Netto LES, Sathian MB, Konrad DB, Covey DF, Michalke B, Bommert K, Bargou RC, Garcia-Saez A, Pratt DA, Fedorova M, Trumpp A, Conrad M, Friedmann Angeli JP. 7-Dehydrocholesterol is an endogenous suppressor of ferroptosis. Nature. 2024 Feb;626(7998):401-410.
[10] Liang D, Feng Y, Zandkarimi F, Wang H, Zhang Z, Kim J, Cai Y, Gu W, Stockwell BR, Jiang X. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. 2023 Jun 22;186(13):2748-2764.e22.
[11] Ryan SK, Zelic M, Han Y, Teeple E, Chen L, Sadeghi M, Shankara S, Guo L, Li C, Pontarelli F, Jensen EH, Comer AL, Kumar D, Zhang M, Gans J, Zhang B, Proto JD, Saleh J, Dodge JC, Savova V, Rajpal D, Ofengeim D, Hammond TR. Microglia ferroptosis is regulated by SEC24B and contributes to neurodegeneration. Nat Neurosci. 2023 Jan;26(1):12-26.




![CRISPR synthetic lethal library screening revealed FSP1[5] CRISPR synthetic lethal library screening revealed FSP1[5]](/uploads/allimg/240318/1-24031Q5320O50.png)
![Both the screening using the genome-wide activation library (A)[6] and metabolic gene knockout library (B)[7] found the enriched GCH1 Both the screening using the genome-wide activation library (A)[6] and metabolic gene knockout library (B)[7] found the enriched GCH1](/uploads/allimg/240318/1-24031Q53441Q2.png)
![The genome-wide knockout library revealed the relationship between distal cholesterol synthesis pathway and ferroptosis[8] The genome-wide knockout library revealed the relationship between distal cholesterol synthesis pathway and ferroptosis[8]](/uploads/allimg/240318/1-24031Q53GBZ.png)



