Published on: December 20, 2024
Views:
--
Hot!!! CRISPR Library for Membrane Proteins: How to Screen for Potential Targets!

Membrane proteins are distributed across various cell membranes and organelle membranes, accounting for approximately 25% of the human proteome. They play a crucial role in cell signaling, interaction with the external environment, and intercellular communication. Dysregulation of membrane protein expression is essential in the development of tumors, cancer cell proliferation, metastasis, and other processes. Membrane proteins are considered important targets in drug discovery and basic research. CRISPR libraries are widely used for target screening due to their low cost, high throughput, and good reproducibility. So, what potential disease treatment targets can be identified through membrane protein libraries?
Mouse Membrane Protein Activation Library Screening Identifies Lrrn4cl as a Tumor Metastasis Target
Melanoma accounts for about 5% of all skin malignancies but has a high mortality rate of over 70% due to its propensity for metastasis. To identify genes associated with melanoma metastasis, Adams et al. used a mouse membrane protein CRISPR activation library to create a B16-F0 library cell line, which was injected into nude mice via the tail vein. On day 19, lung metastatic tumor cells were collected (Figure 1). NGS sequencing analysis revealed that the Lrrn4cl gene was significantly enriched in the metastatic tumor tissue compared to the control group sgRNA[1] (Table 1).
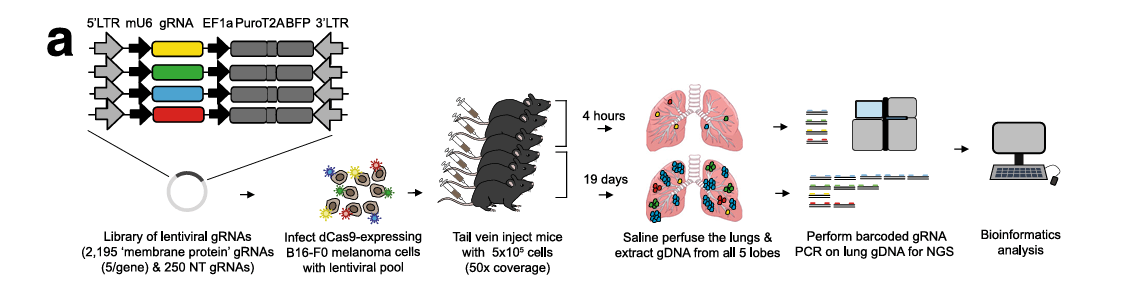
Figure 1: Mouse membrane protein activation library screening identifies genes promoting melanoma lung metastasis[1]
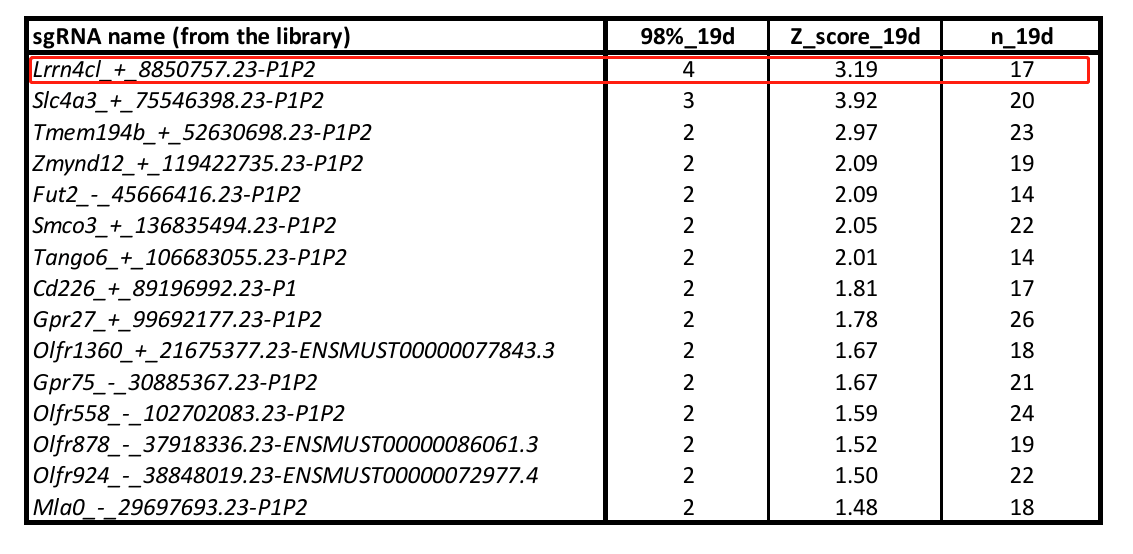
Table 1: sgRNA enrichment analysis in lung tumor tissues[1]
To confirm that Lrrn4cl promotes tumor metastasis, researchers overexpressed the gene in three different mouse melanoma cell lines (Figure 2a) and in three other tumor cell lines (Figure 2b). They found that these cells exhibited significantly enhancing pulmonary metastatic colonisation. Additionally, using the membrane protein activation library to screen E0771-dCas9 cells also showed that Lrrn4cl was the most enriched gene (data not shown), further supporting its importance in tumor cell lung metastasis.
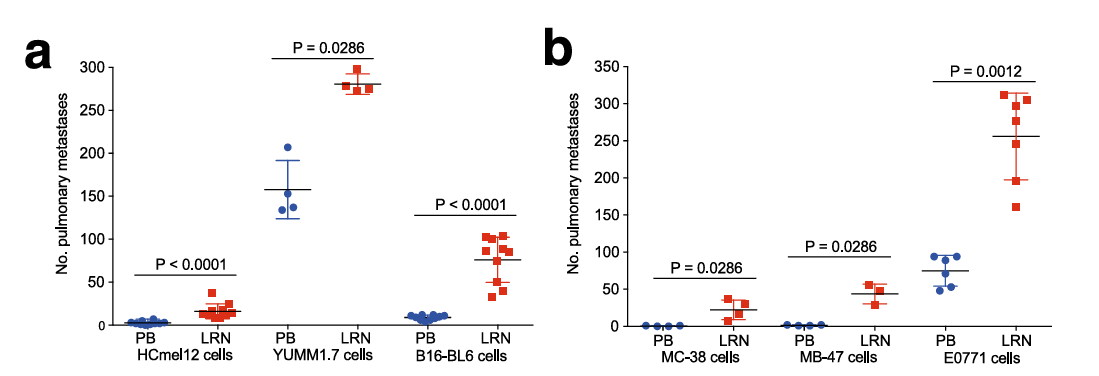
Figure 2: Overexpression of Lrrn4cl in different cell lines enhancing pulmonary metastatic colonisation[1]
Human Membrane Protein Activation Library Screening Identifies TMX2 as a Promoter of HCC Cell Proliferation
Primary liver cancer is the sixth most common cancer type and the third leading cause of cancer-related death. Hepatocellular carcinoma (HCC), the predominant histological type of primary liver cancer, accounts for the majority of liver cancer diagnoses and deaths. Zhang et al. used a human membrane protein CRISPR activation library to infect HCC cells, which were then cultured for successive passages . And then they compared the differences in sgRNA between day 1 and day 14 cells[2] (Figure 3).
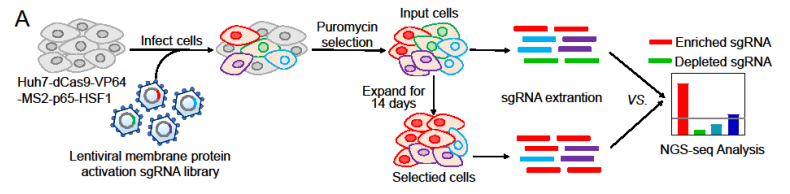
Figure 3: Human membrane protein activation library screening identifies genes promoting HCC cell proliferation[2]
Analysis revealed that the TMX2 gene was significantly enriched in the positive screen. Furthermore, in a separate research using the genome-wide knockout library screening of HCC cells, the TMX2 gene was also significantly enriched in the negative screen[3] (Figure 4). These results suggest that TMX2 plays a crucial role in HCC cell viability.
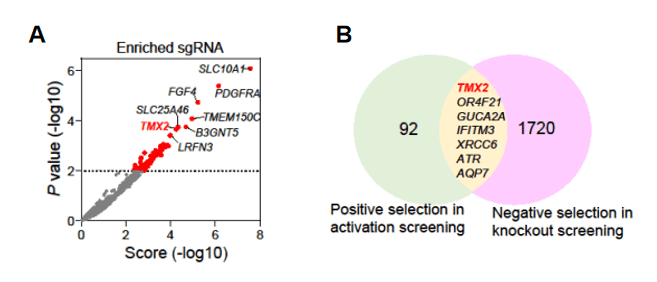
Figure 4: TMX2 gene enrichment in the genome-wide knockout library screening[2]
To confirm the effect of TMX2 on HCC cell viability, the researchers conducted gene knockdown and overexpression experiments in different HCC cell lines. They found that knockdown of TMX2 significantly reduced the clone formation ability of HCC cells, while overexpression notably increased HCC cell proliferation (Figure 5).
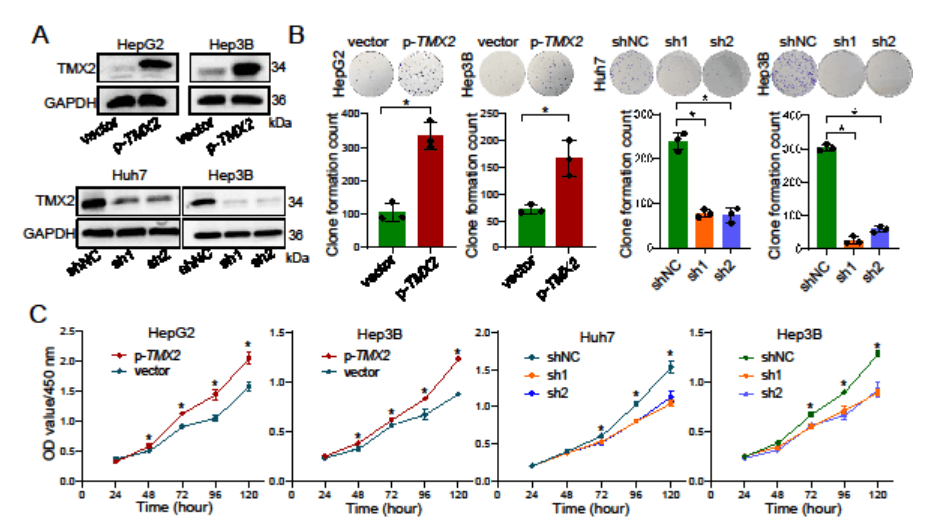
Figure 5: Gene knockdown or overexpression of TMX2 in different HCC cell lines[2]
Further investigation into the mechanism of the effect of TMX2 on HCC cell proliferation revealed that by comparing the difference between TMX2 high expressiong and low expression samples in the TCGA (The Cancer Genome Atlas) database, autophagy-related genes were significantly enriched in the high TMX2 expression samples. RT-qPCR analysis also showed that knockdown of TMX2 led to a significant downregulation of autophagy-related genes (Figure 6).
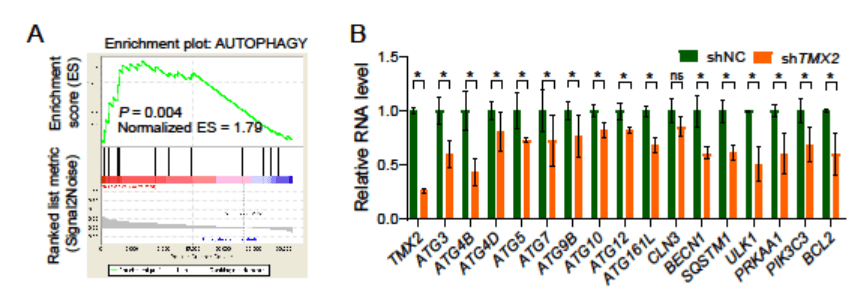
Figure 6: TMX2 regulates autophagy levels in HCC cells[2]
Through co-IP, immunofluorescence, and other experiments, the researchers explored the mechanism of the effect of TMX2 on autophagy and found that TMX2 regulates autophagy by modulating the nuclear-cytoplasmic distribution of the KPNB1 protein (Figure 7).
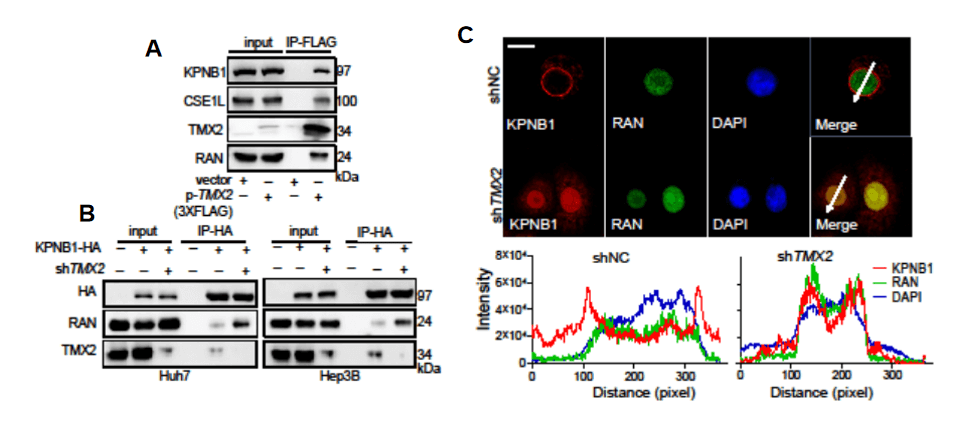
Figure 7: TMX2 regulates autophagy through modulation of KPNB1 nuclear-cytoplasmic distribution[2]
References
[1]van der Weyden L, Harle V, Turner G, Offord V, Iyer V, Droop A, Swiatkowska A, Rabbie R, Campbell AD, Sansom OJ, Pardo M, Choudhary JS, Ferreira I, Tullett M, Arends MJ, Speak AO, Adams DJ. CRISPR activation screen in mice identifies novel membrane proteins enhancing pulmonary metastatic colonisation. Commun Biol. 2021 Mar 23;4(1):395.
[2]Zhang W, Tang Y, Yang P, Chen Y, Xu Z, Qi C, Huang H, Liu R, Qin H, Ke H, Huang C, Xu F, Pang P, Zhao Z, Shan H, Xiao F. TMX2 potentiates cell viability of hepatocellular carcinoma by promoting autophagy and mitophagy. Autophagy. 2024 Oct;20(10):2146-2163.
[3]Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017 Dec;49(12):1779-1784.















