Published on: May 07, 2024
Views:
--
Ubigene's Stable Cells Assist in Revealing the Mechanism of Salmonella Hijacking Macrophages to Breach the Gut-Vascular Barrier
Introduction
Salmonella is an important pathogen that causes diseases such as food poisoning and typhoid in humans and animals. After entering the human body through contaminated food, Salmonella invades the epithelial cells of the small intestine and then spreads to organs such as the liver and spleen. Studies have shown that Salmonella can enter the host's bloodstream within 15 minutes of infection, and its early hematogenous infection is considered a key pathway for systemic spread[1]. Despite the barrier function of the intestinal-blood vessel barrier, Salmonella can quickly breach this barrier and disseminate into the bloodstream and organs[2]. The mechanism of this process remains incompletely understood.
Abstract
Recently, the research teams of Bingqing Li and Qixin Chen from Shandong First Medical University, China, published an article titled “Salmonella manipulates macrophage migration to penetrate the gut-vascular barrier by SteC-mediated MLC activation” on The
EMBO Journal. In this study, the teams used the RAW264.7 cell lines stably overexpressing pSteC, pSteC K256H, and GFP constructed by Ubigene to reveal the early mechanism of Salmonella invading the bloodstream: Salmonella phosphorylates the host's myosin light chain (MLC, Myl12a) through effector protein SteC, hijacking macrophages to penetrate the gut-vascular barrier and then spread into the bloodstream. Notably, SteC exhibits a unique dimeric conformation distinct from known kinases and can utilize four nucleotides (ATP, UTP, GTP, CTP) to catalyze protein phosphorylation, giving Salmonella a survival advantage in the ATP-depleted host environment.
Research Methods and Results
To
investigate the early entry of Salmonella into the bloodstream, the researchers first created a GFP-labeled
Salmonella-infected animal model. Immunofluorescence staining of the mouse intestines within 1
hour post-infection revealed that Salmonella had already accumulated around the blood vessels within 30 minutes of
infection, specifically within CX3CR1+ macrophages. Further analysis of the cell types in the mouse blood
after infection showed that GFP-labeled Salmonella mainly existed in CX3CR1+ macrophages, indicating their
role in breaking the vascular barrier and entering the bloodstream for infection.
The researchers found that macrophage motility and invasion capability significantly increased after Salmonella
infection. However, Salmonella treated with lipopolysaccharide (LPS) or heat did not show enhanced macrophage
motility. Further investigations revealed that Salmonella's activation of macrophage motility and invasion depend on
the actin cytoskeleton, independent of the tubulin cytoskeleton. By constructing effector protein knockout
Salmonella strains involved in cytoskeleton regulation, the researchers identified SteC as the key regulator.
Using BMDM and RAW264.7 cell lines, they verified the effect of Salmonella effector protein SteC on macrophage
motility and invasion.
SteC is the only kinase effector protein secreted by Salmonella's T3SS secretion system. Compared to known kinases, SteC has low homology and there is no similar protein structure to SteC in the PDB. Studies have shown that SteC triggers actin reorganization through the phosphorylation of MEK in fibroblasts, aiding the survival of Salmonella within host cells[3]. Through the stable RAW264.7 cell lines constructed by Ubigene, which respectively expressed pSteC, pSteC K256H, and GFP, the researchers found that the MEK-ERK pathway in macrophages was suppressed after Salmonella infection, suggesting that SteC might regulate macrophage motility through a novel pathway.
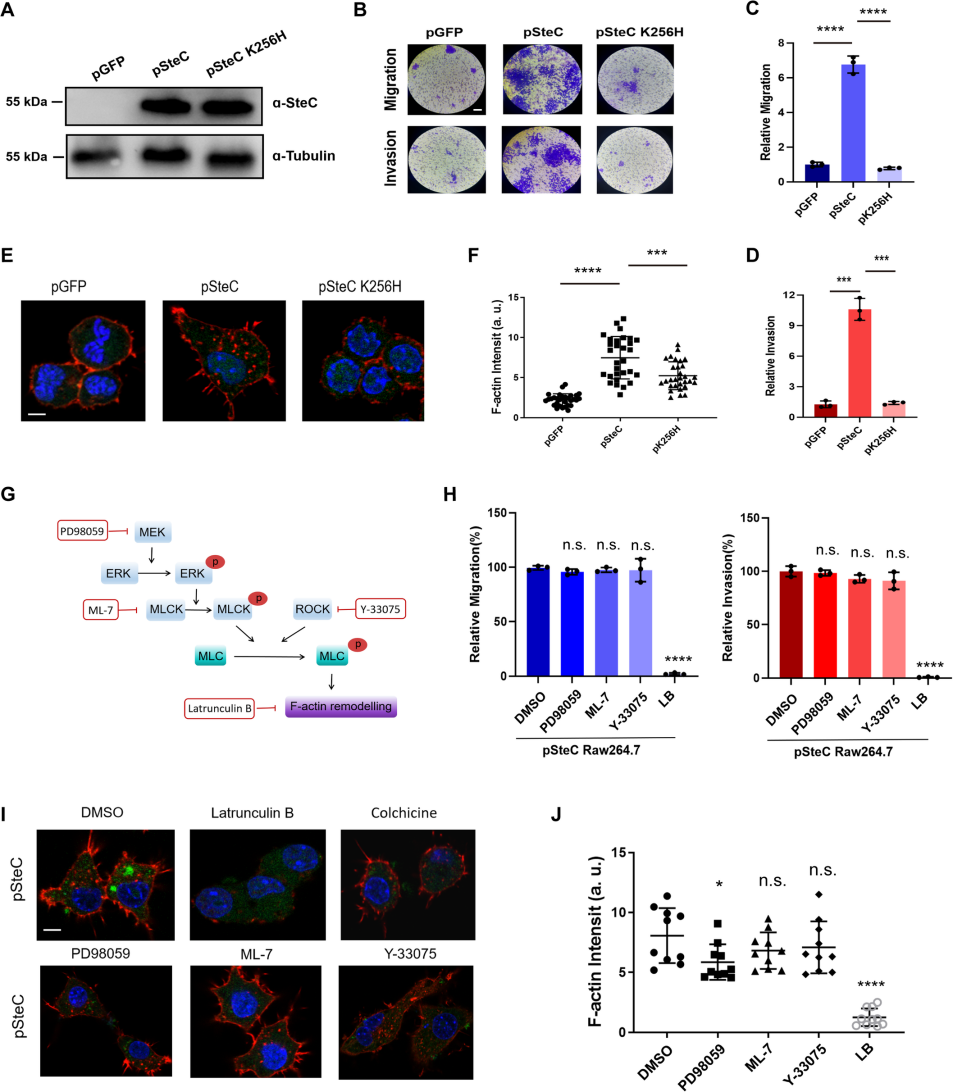
Figure 1. SteC induces actin rearrangement and the migration/invasion properties of macrophages in a MEK/ERK/MLCK pathway-independent manner
To identify the target of SteC in macrophages, the researchers constructed a cDNA library of RAW264.7 macrophages and used SteC as bait for yeast two-hybrid screening, eventually determining that myosin regulatory light chain (MLC, Myl12a) is one of the major targets of SteC in host cells. Further research revealed that SteC can catalyze the activation of Myl12a phosphorylation at Thr19 and Ser20 in vitro and in vivo, independently of common MLC activation pathways in host such MEK-ERK-MLCK and ROCK pathways. In vivo experiments showed that Myl12a phosphorylation dependent on SteC appeared in macrophages within 15 minutes of Salmonella infection. Biochemical analysis revealed that SteC is a unique kinase that can utilize ATP, UTP, CTP, and GTP as phosphate donors for Myl12a phosphorylation. Crystal structure analysis of SteC revealed a unique dimer-mediated kinase activation center conformation, distinct from all reported kinase structures. SteC recruits Myl12a protein by simulating the Myl12a substrate through the long helix structure in front of its kinase domain. In vivo experiments demonstrated that Salmonella lacking steC lost the ability to drive macrophages to penetrate the vascular barrier and cause disseminated multi-organ infection.
Conclusion
In summary, this study reveals a novel mechanism of pathogen hijacking of host cell functions: Salmonella secretes the effector protein SteC into macrophages, phosphorylating and activating the myosin regulatory light chain Myl12a, resulting in actin rearrangement in macrophages and manipulation of macrophage motility and invasion capability. This allows Salmonella to use macrophages to enter the bloodstream and subsequently disseminate to various target organs to cause multi-organ infection (Figure 2).
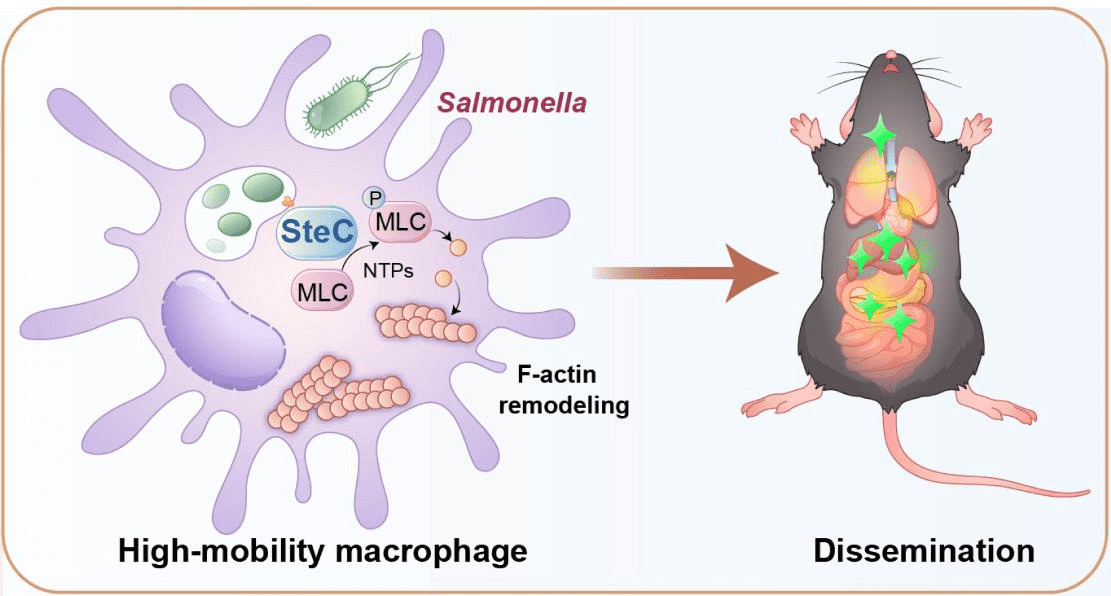
Figure 2. Mechanism model of Salmonella SteC regulating macrophage motility and in vivo dissemination
References
[1]Vazquez-Torres, A., Jones-Carson, J., Bäumler, A. J., Falkow, S., Valdivia, R.,
Brown, W., ... & Fang, F. C. (1999). Extraintestinal dissemination of Salmonella by CD18-expressing
phagocytes. Nature, 401(6755), 804-808.
[2]Spadoni, I., Zagato, E., Bertocchi, A., Paolinelli, R., Hot, E., Di Sabatino, A.,
... & Rescigno, M. (2015). A gut-vascular barrier controls the systemic dissemination of bacteria. Science,
350(6262), 830-834.
[3]Odendall, C., Rolhion, N., Förster, A., Poh, J., Lamont, D. J., Liu, M., ... & Holden, D. W. (2012). The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Cell host & microbe, 12(5), 657-668.