NF-κB pathway, an important target of tumor
therapy
Background
In 1986, Sen and Baltimore first identified a new transcription
factor--NF-κB,
which binds to immunoglobulin
enhancers in activated B cells. Since then, NF-κB's research has
been deepened and proved that it has an impact on tumor cell inflammation, proliferation, survival, apoptosis,
angiogenesis, epithelial-mesenchymal transformation, metastasis, stem cell, metabolism
and therapeutic drug resistance (Fig. 1). Virus infection, necrotic cell products, DNA damage, oxidative stress
and pro-inflammatory cytokines can quickly induce NF-κB. It is worth
noting that NF-κB has constitutive activity in malignant tumor cells and tumor microenvironment,
and is rarely activated due to the changes of its related genes. Therefore,
NF-κB has become a great potential for tumor therapy. Let's discuss further about NF-κB and its progress in tumor treatment.
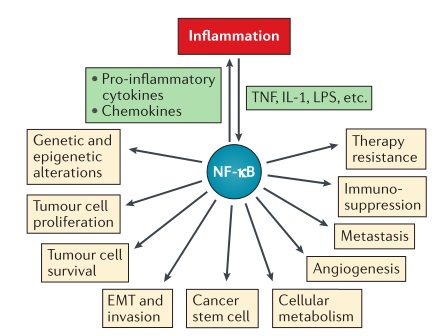
Fig. 1 Roles of NF-κB in tumor
NF-κB signal pathway
NF-κB is an important family of transcription
factors (Rel proteins) with structural similarities. This family is composed of NF-κB1
(p50), NF-κB2 (p52), RelA (p65), RelB and c-Rel
(Fig. 2). In lung cancer, NF-κB members have different roles, for example, NF-κB1 (P105) is associated with longer survival, and the
high expression of p52 related genes is associated with poor prognosis.
NF-κB proteins exist in
cytoplasm as inactive complexes bound with κB family inhibitor (IκB). And the activation of
NF-κB mainly occurs through typical (also known as classical) and atypical (also known as
alternative) signaling pathways (Fig. 3). Typical pathways
include proinflammatory cytokines, PAMPs and DAMPs, which bind to
ligands (such as lipopolysaccharide and bacterial
DNA), and antigen receptors through specific receptors and
adapter molecules, such as TNF receptor (TNFR), IL1R receptor and
toll-like receptor, and then phosphorylated IκB kinase β (IKKβ)
to activate the pathway quickly and transiently. Subsequently, phosphorylated IKKβ forming complex IκB with heterodimer P50/p65,
recruiting proteasome to degrade IκB, resulting in the release of effector
P50/RelA and the translocation of
P50/RelA to κB binding site in the nucleus to achieve its
transcriptional activity (Fig. 3). In contrast, the activation of the
atypical pathway, which depends on de novo
synthesis of NF-κB
inducing kinase, is
much slower. It is activated by
a small number of cytokines in the TNF family, resulting in the
formation of p52/RelB heterodimer (Fig. 3).
NF-κB inducing kinase
(NIK) plays a key role in cell signal transduction and regulation. NIK activation leads to IkB kinase α (IKKα) activation, and mediates the phosphorylation of
P100. Phosphorylated P100 mediates ubiquitination, and finally its degradation is promoted by proteasome to
produce p52. Then, p52 translocates into nucleus according to homodimer or heterodimer, where the complex acts
as a transcription factor.
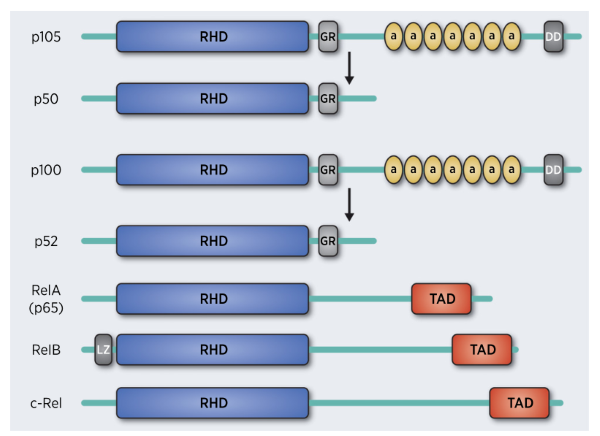
Fig.2 NF-κB family structure
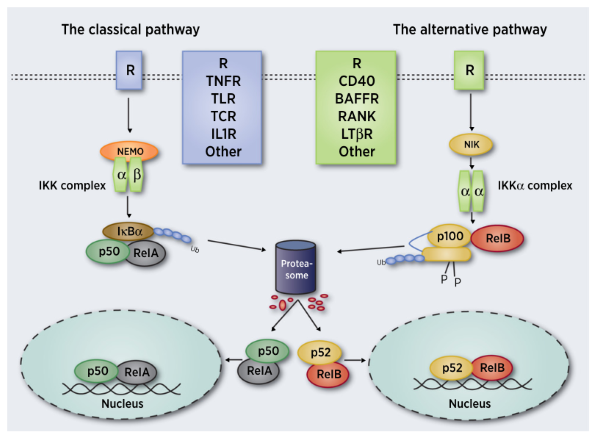
Fig 3. Typical and atypical NF-κB signal pathway
Case studies
1.Identify the role of TRAF3 in RNA and DNA pathways by KO cell line
Some studies have shown that tumor necrosis factor (TNF) receptor related factor 3 (TRAF3)
plays an important role in the activation of IFN mediated by RNA pathway. Mechanistically, TRAF3 is related to
the adapter IPS-1 in the RNA pathway, and the driving signal is activated to the TBK1-IRF3 signal axis. Kislay
et al used mouse embryonic fibroblast cell lines (MEF) of Traf2−/−, Sting−/−, Chuk−/−, Tbk1−/−, Birc2−/−Birc3−/−) to prove that TRAF3, as a negative regulator of IFN reaction activation mediated by
DNA virus or DNA species, plays a direct opposite role in DNA pathway. TRAF3 inhibits nuclear factor
(NF)-κB inducing kinase
(NIK, also known as MAP3K14) leads to impaired IFN activation in DNA pathway and
enhanced IFN response in RNA pathway. NIK binds to the DNA pathway ligand STING
through another mechanism independent of NF-κB pathway, to enhance its
activation. Overall, the data describe a unique signal module (TRAF3/
NIK), which reversely regulates the same signal output (IFN) in a way dependent on signal input (nucleic
acid type), reveals a new regulatory mechanism between RNA and DNA pathway, and provides a conceptual
framework for developing potential therapeutic applications for specific viral infections.
Gene knockout cell lines play a key role in the study of cell
regulation mechanism. Ubigene KO cell bank has nearly 2000
kinds of cell lines in stock, including knockout of TRAF2, Sting, Chuk and other genes. Please visit to learn more>>
2.Demonstrate the switch like
response of feedback interaction between IRF3, NF-κB and STAT pathway to poly (I:
C) by KO cell lines
Maciej et al also used the knockout cell lines of MEFs
(RelA–/–, Stat1–/–) and
PMEFs (Tlr3–/–), and proved that the feedback interaction between
IRF3, NF-κB and STAT pathways
resulted in a switch-like response to the viral analog poly(I: C), rather than a pulse
like response to bacterial LPS. Poly(I: C) activates IRF3 and NF-κB,
which is a necessary condition for inducing IFNβ expression. The autocrine factor
IFNβ initiates JAK/STAT mediated positive feedback stabilization of nuclear IRF3 and NF-κB in first responder cells. In contrast, paracrine
IFNβ up-regulates the positive feedback components RIG-I, PKR and
OAS1A through JAK/STAT mediated positive feedback pathway, which makes the second reactive cells sensitive.
Among these sensitive cells, the "survival or death" determination period after poly(I: C) exposure is short -
they quickly produce antiviral response and lead to apoptosis. In the process of cell population limiting
pathogen transmission, the interrelated positive feedback and feedforward signals are the key to coordinate the
determination of cell destiny.
3.Using KO hESCs to study the regulatory
mechanism
of RelA on human vascular disease and
aging
Blood vessels transport nutrients and oxygen to the whole body through blood. The disorder of
blood vessels will damage the function of vascular cells, resulting in serious cardiovascular diseases such as
myocardial infarction, atherosclerosis and stroke. The activation of constitutive NF-κB in vascular
cells can lead to chronic vascular inflammation, resulting in a variety of cardiovascular diseases. In order to
solve the regulatory mechanism of NF-κB on human vascular cells and guide the treatment of
cardiovascular diseases, the research team of the national geriatric disease clinical research center of Xuanwu
Hospital of Capital Medical University used CRISPR/Cas9 mediated
gene-editing technology to obtain human embryonic stem cells (hESCs) knockout of
RelA and IκBα and differentiate them into various types of vascular cells.
The results of multidimensional phenotypic evaluation and transcriptome analysis showed that the
lack of RelA affected vascular cells by regulating inflammation, survival, angiogenesis, cell
differentiation and cell-specific manner of extracellular matrix tissue under basic conditions,
and RelA could protect vascular cells
against apoptosis and regulate the response of vascular inflammation to tumor necrosis factor α (TNFα) stimulation. Finally,
the gene expression pattern in IκBα knockout vascular cells was further evaluated, which
proved that the role of IκBα was largely independent of RelA signaling pathway.
All in all, NF-κB/RelA has a
protective effect in regulating human vascular homeostasis. At present, in the process of vascular inflammation,
inhibiting NF-κB has provided a promising target for the treatment of various cardiovascular
diseases. And various inhibitors targeting NF-κB pathway have been developed to expand the choice
of clinical application. In view of the cell type specificity of NF-κB regulation, it is proved
that high targeted therapy by inhibiting NF-κB activity is important in the treatment of vascular
inflammatory diseases. At the same time, the establishment of human vascular transcriptome is of great
significance for the discovery of new therapeutic targets.
In the above studies, the CRISPR/Cas9 mediated KO cell lines are indispensable. The targeted KO cell line can not only
quickly verify the positive and negative effects of a gene in the signal pathway on disease treatment, but also
more intuitively and deeply reveal the action mechanism of the signal pathway in cells, so as to provide
guidance for disease treatment. In addition, combined with next generation sequencing, KO cell line is conducive to the discovery of new targets in disease treatment. Therefore, the rapid acquisition of knockout cell lines can accelerate people's comprehensive
understanding of signal pathways including NF-κB, so as to further guide cancer
treatment.
In order to assist
researchers in the in-depth study of major signal pathways, Ubigene has constructed a gene knockout cell
bank with nearly 800 key genes for 8 popular signal pathways, including the NF-κB
signal pathway, such as TRAF3, MAP3K14, and RELA gene knockout cell lines mentioned in this article. As low
as $1980, which takes only one week to deliver! Click here to learn more > >
Reference:
[1]. Dimitrakopoulos, F.D., et al., The Fire Within: NF-κB Involvement in Non–Small Cell Lung
Cancer. Cancer Research, 2020. 80(19): p. 4025-4036.
[2]. Parvatiyar, K., et al., A TRAF3-NIK module differentially regulates DNA vs RNA pathways in
innate immune signaling. Nat Commun, 2018. 9(1): p. 2770.
[3]. Czerkies, M., et al., Cell fate in antiviral response arises in the crosstalk of IRF, NF-κB
and JAK/STAT pathways. Nature Communications, 2018. 9(1).
[4]. Sun, S., The non-canonical NF-κB pathway in immunity and inflammation. Nature Reviews
Immunology, 2017. 17(9): p. 545-558.
[5]. Taniguchi, K. and M. Karin, NF-kappaB, inflammation, immunity and cancer: coming of age. Nat
Rev Immunol, 2018. 18(5): p. 309-324.
[6]. Wang, P., et al., CRISPR/Cas9-mediated gene knockout reveals a guardian role of NF-κB/RelA in
maintaining the homeostasis of human vascular cells. Protein & Cell, 2018. 9(11): p. 945-965.