“Genetic Scissors” CRISPR/Cas9 assists immunotherapy research for hepatocellular carcinoma (HCC)
Worldwide, liver cancer is the sixth most common cancer and the third leading cause of cancer death. It is most common in sub-Saharan Africa and parts of eastern Asia, including China, Hong Kong, and Taiwan. Depending on the type of cell that the cancer started in, there are 5 main types of liver cancer, which are hepatocellular carcinoma (HCC), fibrolamellar carcinoma, bile duct cancer (cholangiocarcinoma), angiosarcoma, and hepatoblastoma. The most common type is hepatocellular carcinoma (HCC).
Immunotherapy has become a significant part of comprehensive treatment of hepatocellular carcinoma. Immunotherapy has been shown to be effective in patients with early HCC, late HCC or HCC recurrence after liver transplantation. There are different types of targeted cancer drugs and immunotherapies. Researchers are studying the best way of having these drugs. And if it’s best to have them with other treatments for liver cancer. These drugs include:| Immunotherapy and targeted cancer drugs | Target |
|---|
| Nivolumab | PD-1 (PDCD1) |
| Pembrolizumab | PD-1 (PDCD1) |
| Cabozantinib | PD-L1 (CD274) |
| Ramucirumab | VEGFR (KDR) |
| HMBD-001 | HER3 (ERBB3) |
| A2AFP | AFP |
Ubigene has 3000+ in-stock KO cell lines, covering a variety of drug target genes, including aforementioned PDCD1, CD274, KDR, and ERBB3. Click here to search our in-stock KO cell lines>>
Application of CRISPR/Cas9 in liver cancer models
Cancer involves multiple point mutations, translocations, and chromosomal losses and gains, so it is challenging to identify driver mutations in cancer. Therefore, to study liver cancer more comprehensively and precisely, it is necessary to model the development and progression of liver cancer and develop liver cancer models with human disease characteristics. CRISPR/Cas9 accelerates this process due to its efficient and straightforward gene-editing capability. Cancer models can be roughly divided into two categories: in vitro and in vivo. Common in vitro models are hepatoma related cell lines. A cancer model with similar biological characteristics to that of a tumor can be constructed by knocking out specific genes using CRISPR/Cas9 technology to simulate the mutation observed during the occurrence and development of liver cancer. Commonly applied in mice, in vivo cancer models are constructed through the induction of oncogenic mutations or chromosomal rearrangements by delivering the CRISPR/Cas9 system to specific organs or tissues in mice.
Hepatoma cell lines are difficult to culture and perform gene-editing because of their low transfection efficiency and difficulty in single-cell cloning. Relying on Ubigene's 12 years of gene-editing experience and the exclusive CRISPR-UTM technology, the efficiency is 10 times higher than traditional methods, making gene-editing in hepatoma cell cells (such as Hep-G2 and Huh-7) easier. Now Ubigene is offering new semester promotion, custom Hep-G2 KO cell line as low as $3780!
If you need to build a mouse model for liver cancer research, Ubigene can provide high-purity and high titer lentivirus and Adeno-associated virus for vivo experiments, starting from $559, deliver as fast as 3 weeks!Contact us to get your exclusive research model construction strategy>>
Case study
Uncover the complex relationship between SNORD17 and p53 network in HCC
Tumor suppressor p53 is activated in response to DNA damage or abnormal activation of oncogenes. It induces a variety of cellular processes, including cell cycle arrest, apoptosis and aging. Studies have shown that p53 plays a key role in the development of HCC. Many non-coding RNAs play an important role in p53 signaling network, including miRNA and lncRNA. However, the role of small nucleolar RNAs (snoRNAs) in p53 regulatory network is not clear. To uncover this mechanism, researchers from the Hepatic Surgery Center of Tongji Hospital edited small nucleolar RNA SNORD17 with CRISPR/Cas9 and revealed the regulatory role of SNORD17 and p53 pathway in hepatocellular carcinoma, which provides a new potential target for the treatment of HCC.
In order to further study the role of SNORD17 in hepatocellular carcinoma, researchers utilized SNORD17 knockout Hep G2 cell line (constructed by Ubigene) and SNORD17 knockdown SK-Hep1 cell line (Fig. 1A). In HCC cell lines, knockout and knockdown of SNORD17 gene can significantly inhibit cell proliferation, clone formation and G1/S phase transition (Fig. 1 B-E). Flow cytometry showed that the knockout and knockdown of SNORD17 gene significantly increased the apoptosis of hepatoma cells compared with the control group (Fig. 1F). To study the role of SNORD17 in the growth of HCC, a subcutaneous xenograft tumor model was established by HepG2 WT cells and HepG2-SNORD17-/- knockout cells. The results showed that knockout of SNORD17 in HepG2 significantly reduced the volume and weight of orthotopic liver tumors compared with the control group (Fig. 1G).
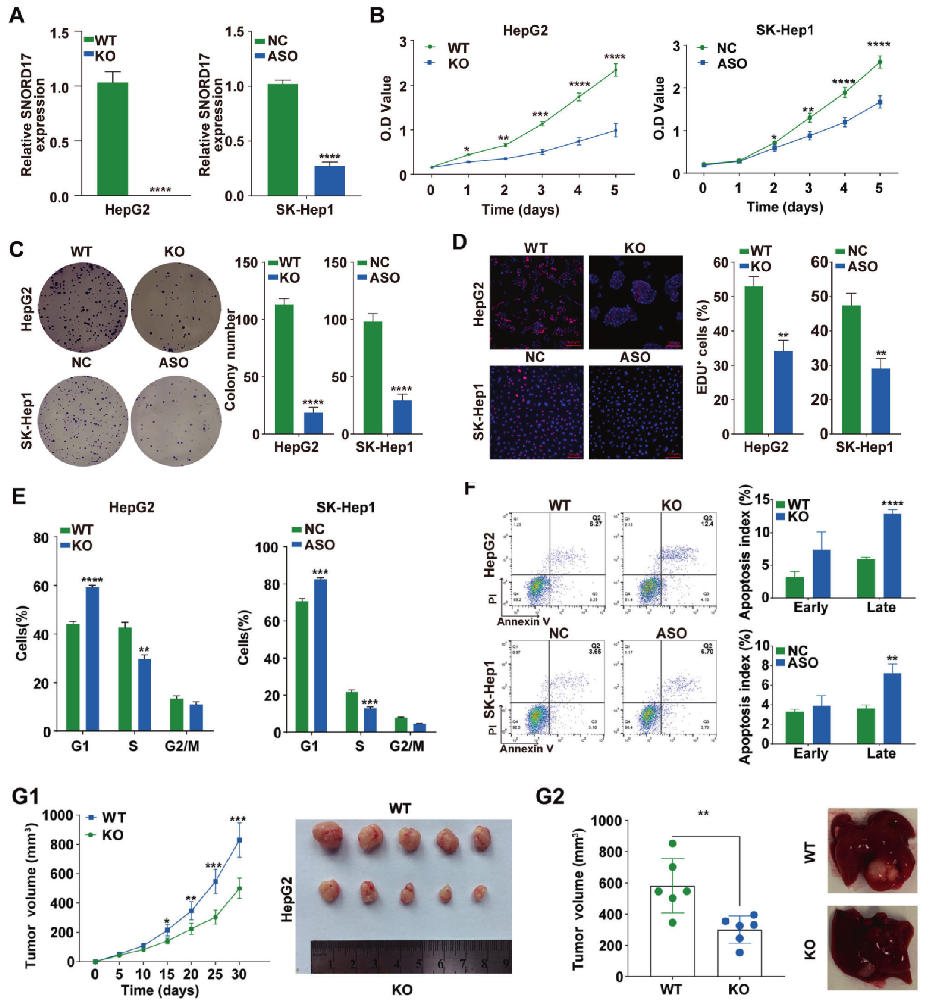
Fig. 1
In order to explore the molecular mechanism of the development of liver cancer promted by SNORD17, the researchers found that p53 can affect the growth of HCC cell line by regulating the expression of SNORD17 through mRNA expression profile, Western Blot, luciferase reporter gene detection, proliferation, and clone formation experiments.
In conclusion, SNORD17 binds to MYBBP1A and inhibits its transfer to nucleoplasm, so as to inhibit p53 acetylation regulated by acetyltransferase p300. SNORD17 regulates the stability and activity of p53 in HCC, and is expected to become a new target for HCC therapy. At the same time, this study also further confirmed the key role of snoRNAs in tumorigenesis and can be used as prognostic markers of various cancers, which could be a reference for subsequent cancer treatment research. Click to view the case details >>
CRISPR/Cas9-mediated knockout of NSD1 suppresses the HCC development via the NSD1/H3/Wnt10b signaling pathway
Recent evidence has indicated that somatic dysregulation of the nuclear receptor binding SET domain-containing protein 1 (NSD1) is associated with the tumorigenesis in HCC, suggesting that NSD1 may serve as a prognostic target for this malignant tumor. In order to investigate how NSD1 regulated HCC progression via regulation of the Wnt/β-catenin signaling pathway, a NSD1 knockout cell line was constructed by CRISPR/Cas9 genomic editing system by Zhang et al., which was investigated in a series of assays such as HCC cell proliferation, migration and invasion, followed by the investigation into NSD1 regulation on histone H3, Wnt10b and Wnt/β-catenin signaling pathway via ChIP.
The results showed that knockout of NSD1 inhibited the proliferation, migration and invasion abilities of HCC cells. CRISPR/Cas9-mediated knockout of NSD1 promoted methylation of H3K27me3 and reduced methylation of H3K36me2, which inhibited Wnt10b expression. The results thereby indicated an inactivation of the Wnt/β-catenin signaling pathway suppressed cell proliferation, migration and invasion in HCC.
Intrinsic activation of β-catenin signaling by CRISPR/Cas9-mediated exon skipping contributes to immune evasion in HCC
Through the comprehensive analysis of clinical samples, the molecular and immunological classification of hepatocellular carcinoma (HCC) was identified by recent studies, and the CTNNB1 (β-catenin)-mutated subtype displays distinctive characteristics of immunosuppressive tumor microenvironment. To clarify the molecular mechanisms, Masafumi Akasu et al. newly developed a highly efficient multiplex CRISPR/Cas9-based genome engineering system for exon skipping by modifying the lentiGuide-Puro plasmid (Nat Method), originally provided by Feng Zhang’s laboratory. By using this newly developed multiplex CRISPR/Cas9-based genome engineering system, human and mouse HCC cell lines with exon 3 skipping of β-catenin were established, which promoted nuclear translocation and activated the Wnt/β-catenin signaling pathway. In β-catenin signaling activating HCC cells, gene set enrichment analysis indicated the downregulation of immune-associated gene sets. By comparing and analysing the gene expression profiles between HCC cell lines harboring wild-type and exon 3 skipping β-catenin, it was elucidated that the expression levels of four cytokines were commonly decreased in human and mouse β-catenin-mutated HCC cell lines. Public exome and transcriptome data of 373 human HCC samples showed significant downregulation of two candidate cytokine genes, CCL20 and CXCL2, in HCC tumors with β-catenin hotspot mutations. T cell killing assays and immunohistochemical analysis of grafted tumor tissues demonstrated that the mouse Ctnnb1Δex3 HCC cells evaded immunosurveillance. To summarise, this study discovered that cytokine regulated by β-catenin signaling activation could contribute to immune evasion, and provided novel insights into cancer immunotherapy for the β-catenin-mutated HCC subtype.
Based on the traditional CRISPR/Cas9 technology, Ubigene has developed exclusive CRISPR-U™ technology with higher gene-editing efficiency. And Ubigene has established a KO cell line bank containing over 3000 in-stock KO cell lines, which includes the relevant KO cell lines mentioned above, such as CTNNB1 KO A549 cell line, CTNNB1 KO HEK293 cell line, WNT10B KO A549 cell line, WNT10B KO HEK293 cell line, SNORD17 KO Hep G2 cell line, etc. And Ubigene has screened nearly 800 key genes from 8 popular signaling pathways, including NF-kB, Hedgehog, JAK-STAT, MAPK, Notch, PI3K-Akt, TGF-beta, and Wnt, to construct KO cell lines. Now only $1980 for a in-stock KO cell line, delivery in 1 week. Our aim is to “make genome editing easier!”







