ERRα/LTK/EMSY The new targets for lung cancer study - Promote Precision Medicine further
Cancer has always been one of the most serious public health events around the world, and it also affects individuals' normal life and work. Due to the lag in statistics, in the 2016 statistical data (as shown in Figure 1), in China, lung cancer had the highest incidence of cases among 4.06 million cancer patients, reaching 828,100 people. Moreover, lung cancer is currently the leading cause of cancer death worldwide, and the number of patients is increasing every year. Therefore, lung cancer is a key research object in the medical field, and targeted therapy has become an indispensable technical means in clinical research. With the continuous advancement of targeted precision therapy research, lung cancer has gradually become a clinically controllable disease, which extends the survival time of advanced patients compared to the era of chemotherapy. Based on the characteristics of EGFR, ALK, ROS1 and other targets, related drugs have been produced and applied in clinical treatment. However, due to the high cost of targeted treatment, many patients are hesitant. Therefore, the key issue is whether more target sites can be discovered and applied. Today, three new hot targets in lung cancer research in recent two years will be introduced: Estrogen-related receptors α (ERRα), Leukocyte tyrosine kinase (LTK) and EMSY transcriptional repressor (EMSY).
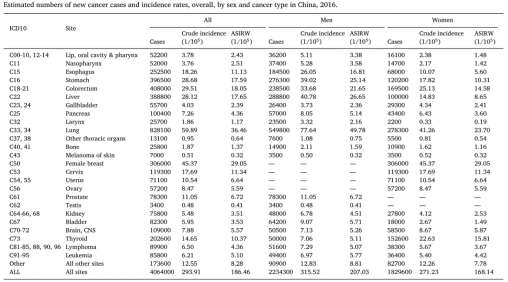
Figure 1. Cancer cases in China in 2016 [1]
Estrogen-related receptors α, ERRα
There are three subtypes of the estrogen-related receptors (ERRs) family: α, β and γ, and this class of proteins belongs to a large family of orphan nuclear receptors, which are involved in muscle differentiation, bone regeneration, immune system regulation, gene regulation, and energy metabolism, etc [2]. The earliest ERRs identified is a steroid receptor screened according to the similarity of DNA binding domain sequence in a cDNA library screening [3]. The basic structure of the ERRs (shown in Figure 2A) consists of an activation function domain 1 (AF1), a DNA binding domain (DBD), a ligand binding domain (LBD), and an activation function domain 2 (AF2). Four domains of the three subtypes of ERRs: α, β and γ has relatively high conservation (as shown in Figure 2B). Normal ERRs are involved in various cellular metabolisms and maintain cellular homeostasis, and a recent review summarizes recent findings that aberrantly expressed ERRs can be the “culprits” of tumors.
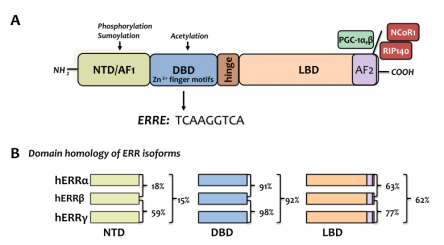
Figure 2. (A) The basic structure of the ERRs family; (B) The homology comparison of the three subtypes of the ERRs family [2]
Among them, ERRα was despite defined as an orphan nuclear receptor at the outset, there were soon relevant studies demonstrating that ERRα has other counterpart ligands to bind to involve in gene regulation, however, it is still defined as an orphan nuclear receptor and has been to date. ERRα is involved in the process of tumorigenesis in both estrogen-dependent and -independent manners. The aberrant expression of ERRα is associated with breast, prostate, non-small cell lung and other cancers [4]. Therefore ERRα is gradually treated as a potential target for cancer therapy. A recent study found that re-expression of the ERRα can increase NSCLC drug resistance by activating the EGFR/Src/Erk/SP1 signaling pathway (as shown in Figure 3), which attenuates the efficacy of EGFR inhibitors EGFR-TKIs [5]. Here, some articles identified ERRα, the downstream protein of the EGFR/Src/Erk/SP1 signaling pathway, has high expression in drug-resistant lung cancer cell lines selected by EGFR inhibitors EGFR-TKIs. Downregulation of cholesterol levels by Lovastatin or the blockade of the pathway by ERRα inhibitor XCT790 can effectively inhibit the growth of drug-resistant lung cancer cells. Also a more recent study in 2023, Avinash et al developed BipotentR, a proteomic approach to successfully predict and validate ERRα drug resistance is the result of the inhibition of two immune pathways: A. cytokine induction, causing pro-inflammatory macrophage polarization; B. stimulation by antigen presentation, which guides CD8+ T cells into the tumor, in addition to energy metabolism pathways [6]. The more well-established ERRα knockout animals and cell models has been described for research [7]. Ubigene provides CRISPR/Cas9 technology for ERRα knockout and stable cell models with ERRα-/- genotype, providing a good platform for discovering other functions of ERRα and the roles played in various types of diseases, such as muscle differentiation, immune system, bone regeneration, and carcinogenesis, among others.
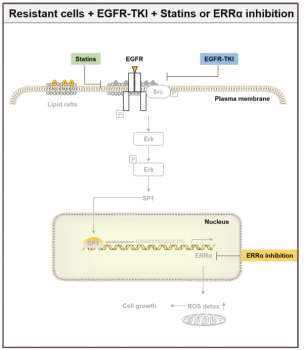
Figure 3. EGFR/Src/Erk/SP1 signaling pathway [5]
Leukocyte tyrosine kinase, LTK
Chromosomal rearrangements resulting in gene fusions are an important oncogenic driver, and the fused genes vary in different tumor types. In a recent article in Nature, Izumi et al found that the CLIP1-LTK fusion protein (shown in Figure 4), but not LTK protein alone, is a cancer-causing factor in lung cancer and accounts for 0.4% of lung cancer cases [8]. Researchers determined the presence of LTK rearrangements in 18% of cancer patient cancer cells by using fluorescent probes with two different fluorescent signals, LTK5’ orange and 3' green, i.e., only one type of 3 'green fluorescence. Exon 16 of CLIP1 (CAP-Gly domain containing linker protein 1) was determined by sequencing to join with exon 11 of LTK to form a novel fusion protein, CLIP1-LTK. Leukocyte receptor tyrosine kinase (LTK) is a member of the ALK/LTK subfamily of the insulin receptor superfamily of RTKs and shares nearly 80% homology with the ALK kinase domain. Inhibitors of ALK, such as Lorlatinib, can therefore also have inhibitory effects on cancer cells positive for the CLIP1-LTK fusion protein (Figure 6). Ubigene provides CRISPR/Cas9 technology for LTK knockout for probing the metabolic pathway changes or resistance mechanism changes compensated for CLIP1-LTK fusion protein null genotypes, or getting different LTK mutant lines obtained by site-directed mutagenesis of LTK based on different clinical data, or exploring other functions in different mutant lines and other mechanisms causing cancer or screening new drugs, etc.
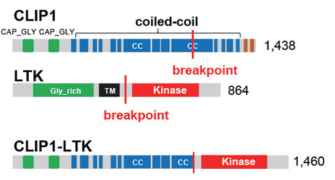
Figure 4. Structure of CLIP protein, LTK protein and CLIP-LTK fusion protein
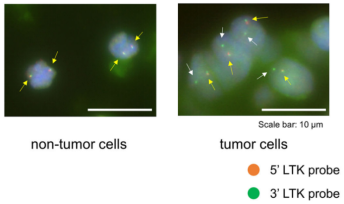
Figure 5. FISH (Fluorescence In Situ Hybridization) experiment of LTK [9]
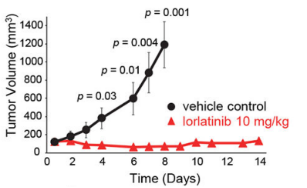
Figure 6. In vivo inhibition of CLIP1-LTK fusion protein positive tumors by Lorlatinib
EMSY transcriptional repressor, EMSY
With the upgrade of scientific research means and the increase of clinical databases, clinical studies have found that EMSY is abnormally highly expressed in both breast and ovarian cancer, thus also gradually becoming an emerging therapeutic target. It was only isolated and confirmed in 2003 that EMSY binds to BRCA2 and BRCA2-like proteins and has functions related to DNA repair and transcriptional regulation [10]. Recently, it was reported that abnormally high expression of EMSY was detected in lung cancer [11]. This study found that non-small cell lung cancers (NSCLCs) harboring KEAP1 mutations are often resistant to immunotherapy and summarized the following pathway modalities (shown in Figure 7). By genetically knocking out the KEAP1, the researchers found that EMSY accumulates and starts to inhibit cellular gene homologous recombination repair and interferon type I immune response, prompting the immune escape of lung cancer cells. It is finally speculated that mutations in KEAP1 (kelch like ECH associated protein 1) in cancer patients result in blockage of the EMSY ubiquitination degradation pathway, which then causes EMSY accumulation (as shown in Figure 8), ultimately leading to lung cancer resistance to immunotherapy. Accumulation of EMSY acts as a double-edged sword, increasing immunotherapy tolerance on the one hand, and resulting in such tolerant lung cancer cells being more sensitive to inhibitors of PARP (poly ADP-ribose polymerase 1) or agonists of STING on the other hand. Like the one studied in the article, Ubigene can generate site-directed knockout of genes (e.g. KEAP1) or site-directed mutagenesis to EMSY for ubiquitination escape according to customers’ need, to activate the pathway and refine other unknown sites in the pathway, helping you to study signaling pathways more time-saving and effort-saving.
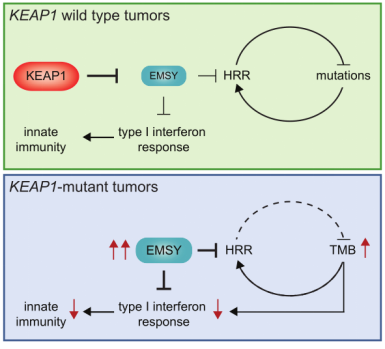
Figure 7. Signaling pathway of KEAP1-EMSY [11]
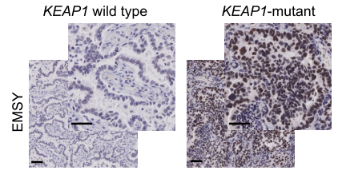
Figure 8. Immunohistochemical maps of tissue samples from KEAP1 wild-type and mutant patients [11]
Summary
Cancer has been a research hotspot in the field of medicine. But there are many proto-oncogenes yet to be discovered, and little is known about the mechanism of action of tumor suppressor genes. Above, three novel therapeutic targets were briefly described in lung cancer: Estrogen-related receptors α (ERRα), Leukocyte tyrosine kinase (LTK) and EMSY transcriptional repressor (EMSY). These three targets were found late and have been little studied, but also gradually being popular targets. The continued discovery of tumor target proteins drives a faster coming era of precision oncology therapeutics and promotes biomarker-oriented treatment paradigms in advanced treatment, and we look forward to more effective treatment modalities in the future.
The aforementioned EGFR, ERRa (ESRRA) and KEAP1 KO cell lines are available in-stock in Ubigene’s KO cell line bank. And the bank also includes RB1, PIK3CB, ERBB2, AKT1, MYC and other hot targets in lung cancer research, as low as $1980 each, deliver in 1 week. Click to check more KO cell products >>
Ubigene also provides express customization as fast as 4 weeks, with comprehensive strategy evaluation and gene risk assessment analysis, helping your experiment follow the right path. Get a customized proposal >>
[1] Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W and He J. Cancer incidence and mortality in China, 2016. Journal of the National Cancer Center 2022; 2 1-9.
[2] Huss JM, Garbacz WG and Xie W. Constitutive activities of estrogen-related receptors: Transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim Biophys Acta 2015; 1852: 1912-1927.
[3] Giguere V, Yang N, Segui P and Evans RM. Identification of a new class of steroid hormone receptors. Nature 1988; 331: 91-94.
[4] Mukherjee TK, Malik P and Hoidal JR. The emerging role of estrogen related receptoralpha in complications of non-small cell lung cancers. Oncol Lett 2021; 21: 258.
[5] Pan Z, Wang K, Wang X, Jia Z, Yang Y, Duan Y, Huang L, Wu ZX, Zhang JY and Ding X. Cholesterol promotes EGFR-TKIs resistance in NSCLC by inducing EGFR/Src/Erk/SP1 signaling-mediated ERRalpha re-expression. Mol Cancer 2022; 21: 77.
[6] Sahu A, Wang X, Munson P, Klomp JPG, Wang X, Gu SS, Han Y, Qian G, Nicol P, Zeng Z, Wang C, Tokheim C, Zhang W, Fu J, Wang J, Nair NU, Rens JAP, Bourajjaj M, Jansen B, Leenders I, Lemmers J, Musters M, van Zanten S, van Zelst L, Worthington J, Liu JS, Juric D, Meyer CA, Oubrie A, Liu XS, Fisher DE and Flaherty KT. Discovery of Targets for Immune-Metabolic Antitumor Drugs Identifies Estrogen-Related Receptor Alpha. Cancer Discov 2023; 13: 672-701.
[7] Mao L, Peng L, Ren X, Chu Y, Nie T, Lin W, Zhao X, Libby A, Xu Y, Chang Y, Lei C, Loomes K, Wang N, Liu J, Levi M, Wu D, Hui X and Ding K. Discovery of JND003 as a New Selective Estrogen-Related Receptor alpha Agonist Alleviating Nonalcoholic Fatty Liver Disease and Insulin Resistance. ACS Bio Med Chem Au 2022; 2: 282-296.
[8] Izumi H, Matsumoto S, Liu J, Tanaka K, Mori S, Hayashi K, Kumagai S, Shibata Y, Hayashida T, Watanabe K, Fukuhara T, Ikeda T, Yoh K, Kato T, Nishino K, Nakamura A, Nakachi I, Kuyama S, Furuya N, Sakakibara-Konishi J, Okamoto I, Taima K, Ebi N, Daga H, Yamasaki A, Kodani M, Udagawa H, Kirita K, Zenke Y, Nosaki K, Sugiyama E, Sakai T, Nakai T, Ishii G, Niho S, Ohtsu A, Kobayashi SS and Goto K. The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer. Nature 2021; 600: 319-323.
[9] The CLIP1-LTK Fusion Is an Oncogenic Driver of NSCLC. Cancer Discov 2022; 12: 286.
[10] Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C and Kouzarides T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 2003; 115: 523-535.
[11] Marzio A, Kurz E, Sahni JM, Di Feo G, Puccini J, Jiang S, Hirsch CA, Arbini AA, Wu WL, Pass HI, Bar-Sagi D, Papagiannakopoulos T and Pagano M. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion. Cell 2022; 185: 169-183 e119.














