Fail to get KO
homozygotes, the reason is that you may knock out the essential gene!
Introduction
Even having rich experience in gene-editing, you may
experience the following cases:
1) The transfection efficiency is high, observing a high coverage of green
fluorescence;
2) gRNA cutting efficiency is high, double peaks at PAM site is observed from
the sequencing of the pool;
But once you
get the sequencing results of the single-cell clones, only WT cells or heterozygotes (KO+WT) is obtained, or
with 3-n deletion. Then you run the experiment again, but still, the same result is obtained. Have you
considered the reason for this scenario?
Knocking out the gene
of interest from cells, and observing the resulting phenotype, is a common strategy used in biological
research to determine gene function, such as for studying cellular signal transduction, regulation of gene
expression, or structural composition. However, there are genes that are essential for cell viability and the knockout of these
genes can cause cell death, and it is likely
that the scenario described in the introduction is the results of knocking out an essential gene therefore unable to get KO homozygotes.
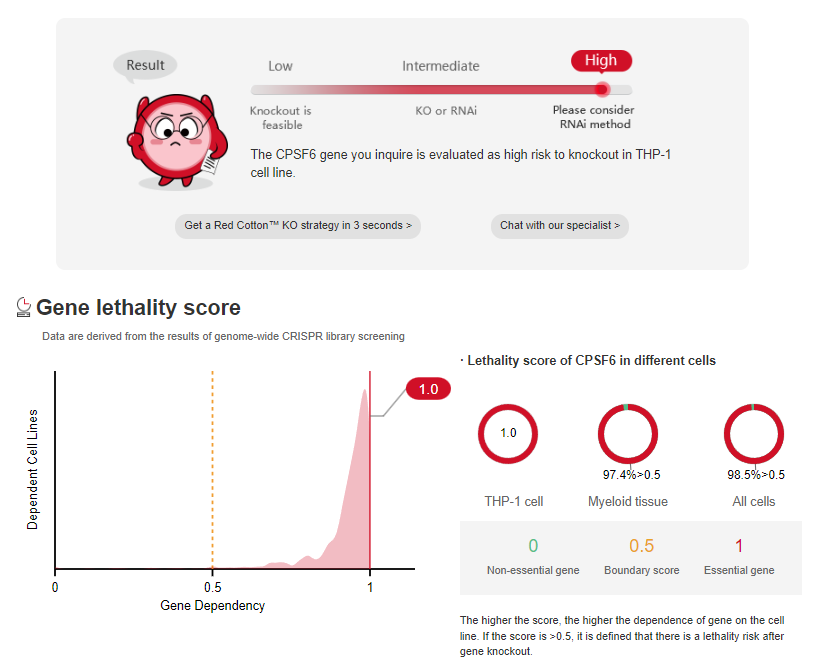
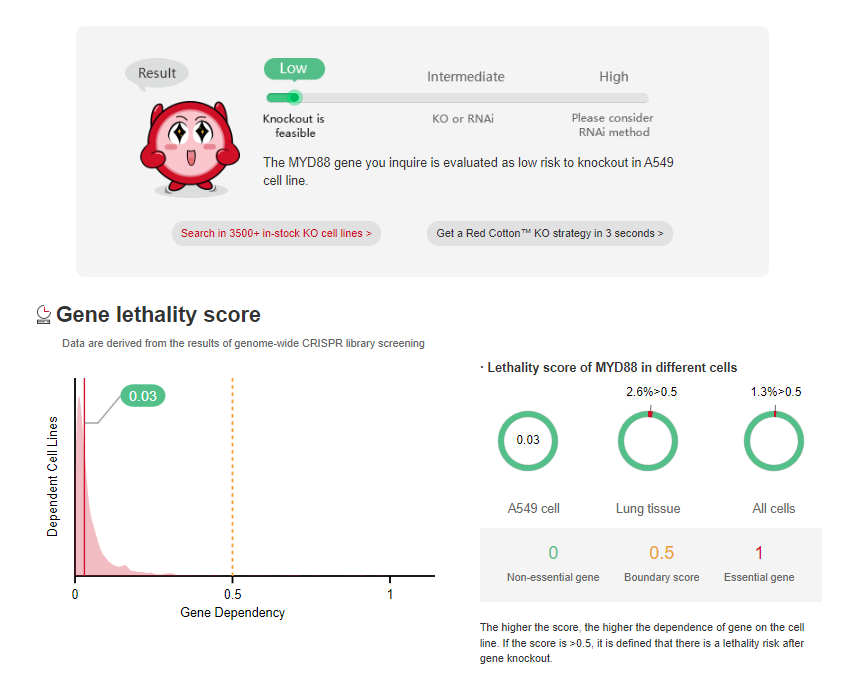
According to the results of
genome-wide screening, essential
genes account for approximately 10% of the total human genes [1]. If
you're not sure if a gene is an essential one for a cell, is there a way to search for it prior to the
experiment? The first thought may be to look up the
literature to see if there are successful knockout cases of the same genes and cells of interest. But after
all, it is time and effort to review a bunch
of articles, and often the
exact same cases with the same gene and cell
line cannot be found. With the wide application of
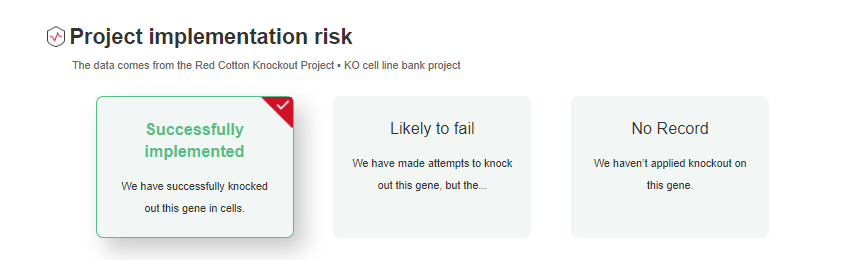
CRISPR library screening, more and more cell genome-wide knockout data are disclosed. Ubigene has gathered nearly 30 million CRISPR library screening data
from 1400 + cell types, allowing you to assess project feasibility (Figure 1&2) and find success cases (Figure 3) simply by entering cell and gene
names. Try it for free >>>
In other words, if you still want to perform the knockout of a high-risk gene, is it no way to achieve it? Of course not, Ubigne has sorted out 3
major solutions for you based on years of experience with gene-editing cell construction.
1) Alternative method to knock down the
gene. If the knockout of the gene leads
to cell death, it is possible to observe whether there are phenotypic changes by repressing the expression
of the gene by RNAi. If the knockdown has a phenotype, generally, the result has a certain value for your
publication.
RNAi case study: downregulation of MTA1 expression by an
RNAi approach can lead to ER α re-expressed in ER-negative breast cancer cell line
MDA-MB-231, the decrease of the protein levels of MMP-9 and CyclinD1, as well as reduced tumor cell invasion
and proliferation. More cells were blocked in G0/G1 phase (P < 0.05). The silencing effect of MTA1 could
effectively inhibit the invasion and proliferation of MDA-MB-231 cells. shRNA interference against MTA1 may
have potential therapeutic utility in human breast cancer [2].
RNAi literature case: down regulation of MTA1 expression
by an RNAi approach can lead to ER α Re expressed in Er negative breast cancer
cell line MDA-MB-231, and decreased the protein levels of MMP-9 and CyclinD1, as well as reduced tumor cell
invasion and proliferation, more cells were blocked in G0 / G1 phase (P < 0.05). The silencing effect of
MTA1 could effectively inhibit the invasion and proliferation of MDA-MB-231 cells. ShRNA interference
against MTA1 may have potential therapeutic utility in human breast cancer [2].
2) Challenge the
probability. If the interference effect
is not good, but the gene is also just lethal with some probability, you can try isolating more clones to
see if you can be fortunate to obtain homozygotes. If no homozygote is obtained but a KO heterozygote or KO
pool, you can also detect the expression of the cells. Sometimes KO heterozygotes also have a phenotype that
is available for publication.
KO heterozygotes case
study: NAT10 is a nucleolar protein containing an acetyltransferase domain and a tRNA
binding domain with histone acetylation activity. NAT10 is involved in
the regulation of human telomerase reverse transcriptase. This gene is an essential gene in the vast
majority of tumor cells and the knockout of this gene leads to cell death. One of the researchers used
CRISPR technology to perform knockout in LoVo cells and the KO heterozygote (NAT10+/-) was obtained and performed with immunofluorescence, Western blotting, and
sequencing validation. Compared to control cells (LoVo), NAT10+/− cells showed reduced MN formation, i.e. downregulation of NAT10 expression can inhibit colorectal cancer progression
[3].
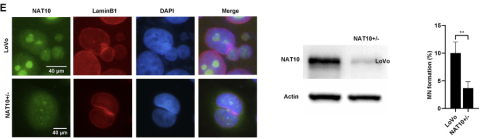
Figure 4. Downregulation of NAT10 expression reduced MN
formation
Here is a supplementary
tip, by adding some special supplements to the media to additionally replenish the nutrients needed for cell
growth, the cells with the essential gene knocked out (that induces lethal) can grow and proliferate
normally. Until the cells expand to sufficient amounts, the special supplements can be withdrawn and the
cells can be used for functional validation experiments. For example, after the knockout of the DHODH in
leukemia cell lines, the cells require ~100µM uridine added to the culture medium to maintain cell growth [4].
3) Adjust strategy to conditional
knockout. As demonstrated by using the tet-off system in combination with CRISPR/Cas9. First to exogenously express the
gene of interest using the tet-off system, followed by knockout of the endogenous gene. Then screen for KO
homozygous clones, and after expansion to a sufficient amount, add dox to switch the expression of the
exogenous gene off, achieving inducible gene knockout [5]. Or by using an Auxin-inducible degron system (AID) in which AID is knocked in to form a fusion protein
replacing endogenous gene expression, then induces the protein degradation by auxin addition, achieving gene
function knockout [6]. In addition, there are strategies to combine CRISPR/Cas9 and cre/loxp systems for
conditional knockout.
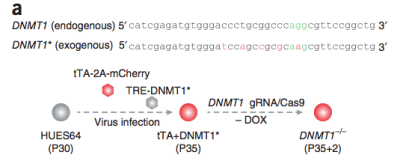
Figure 5.
Conditional knockout strategy 1
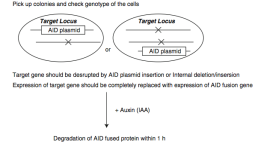
Figure 6.
Conditional knockout strategy 2
The above 3 solutions, each with
advantages and disadvantages. The 1st RNAi alternative method is the easiest to operate, less expensive, and has a
high success rate, but there is a possibility that the interference effect is not good and the knockdown is not
thorough. The second, the approach to increase the number
of clones to be isolated and screen, is appropriate only
for genes with lower lethality, has higher uncertainty, and needs more time and costs.
The third, conditional knockout strategy is the most costly, the tet-off system may also be affected by background expression in some cases, and the KI approach is
more difficult. Overall, the choice of strategies can be made by combining time, funding and specific
genes.
Ubigene’s KO cell bank now is expanded to 3500 + KO cells, covering 1200+
genes from 26 signaling pathways, 1500+ genes from 27 disease types
and 800+ genes from 5 major classes of drug targets, as low as $1980, as fast as 1 week to deliver, click to search > > > Ubigene also offer express customization, as fast as 4 weeks only. With
comprehensive project evaluation and gene dependency assessment, Ubigene can help you achieve the experiment goal in one-stop and more
reliably!
[1]Wang T, Birsoy K, Hughes N W, et al. Identification and
characterization of essential genes in the human genome[J]. Science, 2015, 350(6264): 1096-1101.
[2] Jiang Q, Zhang H, Zhang P. ShRNA-mediated gene silencing of MTA1 influenced
on protein expression of ER alpha, MMP-9, CyclinD1 and invasiveness, proliferation in breast cancer cell
lines MDA-MB-231 and MCF-7 in vitro[J]. Journal of Experimental & Clinical Cancer Research, 2011, 30(1):
1-11.
[3] Cao Y, Yao M, Wu Y, et al. N-acetyltransferase 10 promotes micronuclei
formation to activate the senescence-associated secretory phenotype machinery in colorectal cancer cells[J].
Translational Oncology, 2020, 13(8): 100783.
[4] Sykes D B. The emergence of dihydroorotate dehydrogenase (DHODH) as a
therapeutic target in acute myeloid leukemia[J]. Expert opinion on therapeutic targets, 2018, 22(11):
893-898.
[5] Liao J, Karnik R, Gu H, et al. Targeted disruption of DNMT1, DNMT3A and
DNMT3B in human embryonic stem cells[J]. Nature genetics, 2015, 47(5): 469-478.
[6] Nishimura K, Fukagawa T. An efficient method to generate conditional knockout
cell lines for essential genes by combination of auxin-inducible degron tag and CRISPR/Cas9[J]. Chromosome
Research, 2017, 25: 253-260.