IF=14.4丨Ubigene's Contribution to the Identification and Characterization of Salmonella Typhimurium Mutants

Background
Salmonella is a common intestinal pathogen worldwide, and
Salmonella enterica serovar Typhimurium (
S. Typhimurium) is one of the leading causes of salmonellosis in humans. In recent years,
various dominant strains of this bacterium have emerged, posing serious public health threats. With the advancement of whole-genome sequencing technologies, new methods have been developed for the epidemiological study of
Salmonella.
Summary
Recently, the PLA Center for Disease Control and Prevention published a paper in Nature Communications (IF=14.4) titled “Characterization of a Salmonella enterica serovar Typhimurium lineage with rough colony morphology and multidrug resistance”. This study identified and characterized a rough colony variant of S. Typhimurium, which may have originated from the smooth colony strains of China around 1977. The strain primarily belongs to the L6.5 sub-lineage and carries a specific mutation in the csgD promoter. In this study, the research team utilized a csgD knockout strain constructed by Ubigene to investigate the role of the csgD gene in mediating the morphological transition to the mrdar variant and in biofilm formation.
Figure and Text Overview
1. Characteristics and Epidemiology of the Variant
In this study, 2,212 S. Typhimuriumisolates from China were collected, with 29.6% identified as rough colony variants. These variants were mainly distributed in the eastern, southern, southwestern, and central regions of China, and were isolated from humans, food, and the environment, with the highest proportion of isolates coming from environment. The rough colony variants exhibited rough, membranous, and wrinkled colony morphology, and demonstrated stronger biofilm formation at both 28°C and 37°C compared to smooth colony strains and control strains. Antibiotic susceptibility tests revealed that S. Typhimuriumisolates from China generally showed high levels of antibiotic resistance, with the rough colony variants exhibiting even higher resistance rates to multiple antibiotics compared to the smooth colony strains.
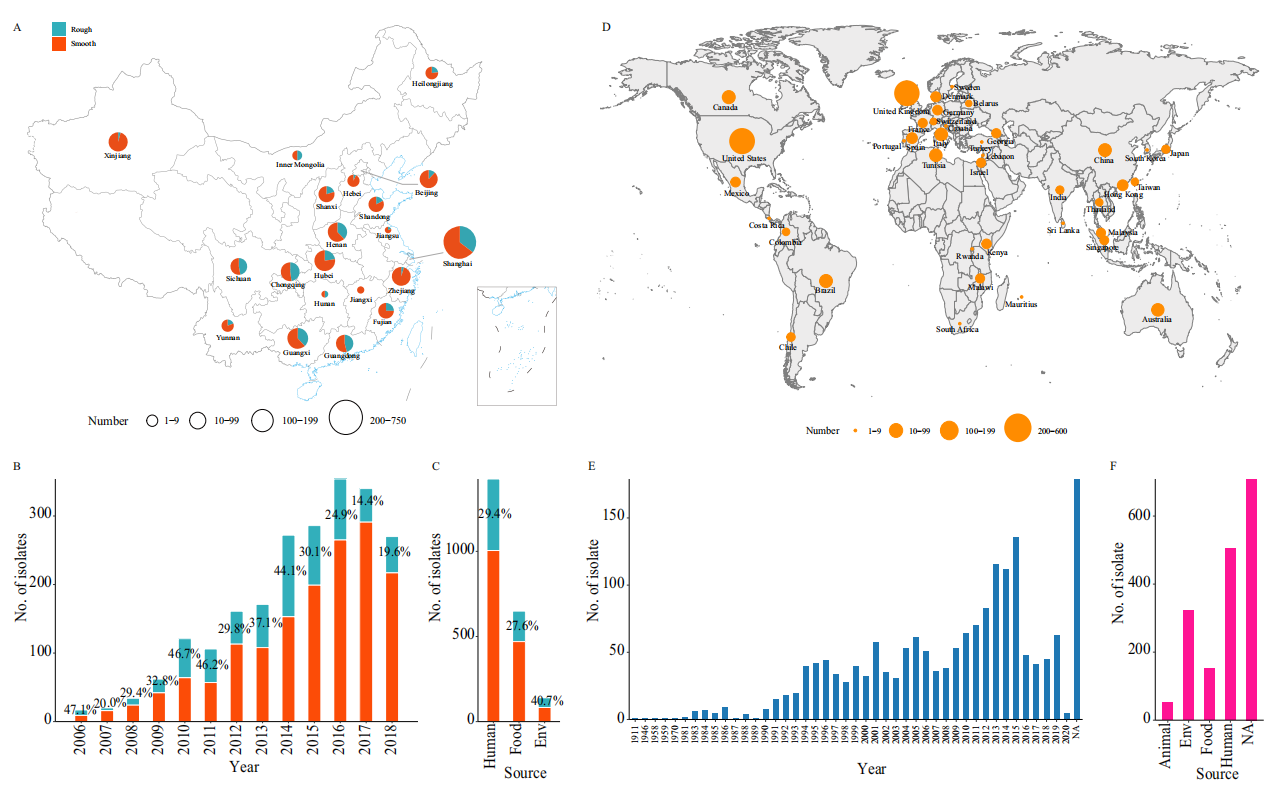
Figure 1: Distribution of Salmonella typhimurium isolates
2 Population Structure and Phylogenetic Analysis
The global S. Typhimurium population can be divided into six different lineages (L1-L6), with isolates from China primarily distributed in lineage L6 (59.0%). Most rough colony variants (93.9%) belong to the sublineage L6.5, which can be traced back to 1977 and is possibly originated from China. It is noteworthy that L6.5 also includes isolates from other countries, such as Europe and North America.
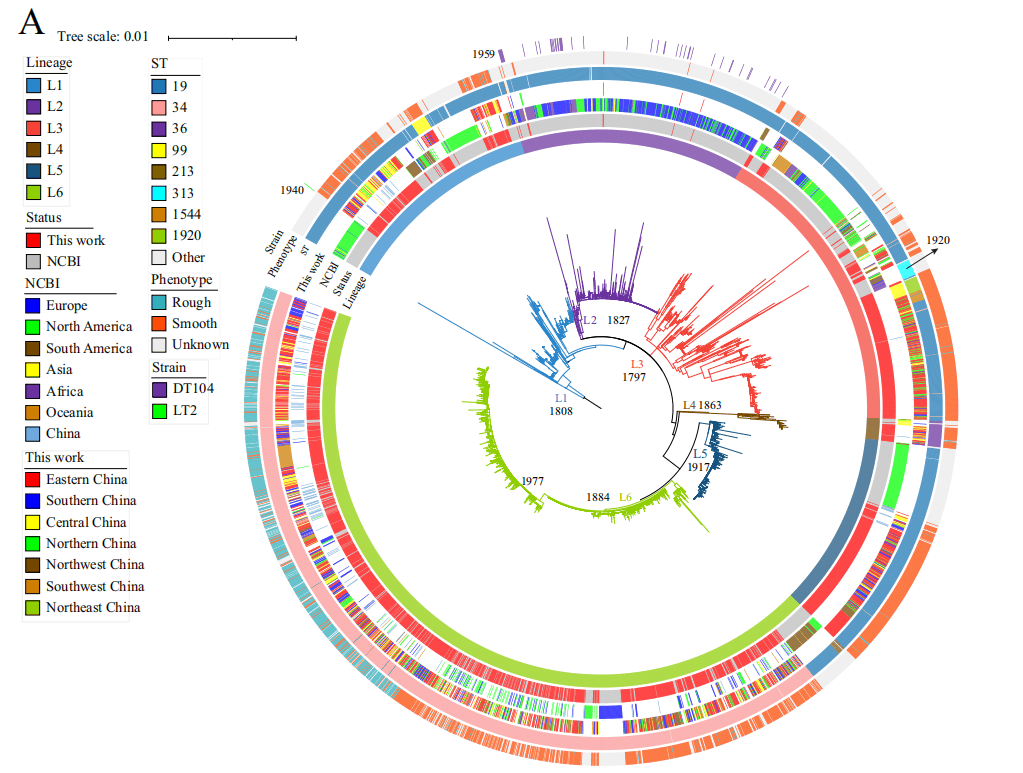
Figure 2: Phylogenetic tree of 3951 S. Typhimurium isolates globally
3 AMR and Virulence Factor Analysis
A total of 152 antimicrobial resistance (AMR) determinants were identified, with rough colony variants carrying more of these determinants. The sublineage L6.5 harbors the greatest number. A total of 265 virulence genes were identified from a virulence factor database. Rough colony variants generally have fewer virulence genes than smooth colony strains, but in lineage L6, they carry more virulence genes than smooth isolates. Sublineage L6.5 mainly carries the fljAB gene.
4 Plasmid Analysis
A total of 47 plasmid replicons were predicted. The IncFII(S) replicon was detected in lineages L1, L2, and L3, while the IncHI2 replicon was found in sublineage L6.5. Two complete plasmids, pSH15G1765-2 and pYY-2016-057-2, were generated using Illumina and Nanopore sequencing data. The plasmid pSH15G1765-2 belongs to IncHI2 and is prevalent in L1, L2, and L3 strains, carrying virulence genes such as pefABCD, spvBCD, and rck, as well as the merTPCA metal resistance gene. The plasmid pYY-2016-057-2 also belongs to IncHI2 and is prevalent in L6.5 strains, carrying 19 resistance genes.
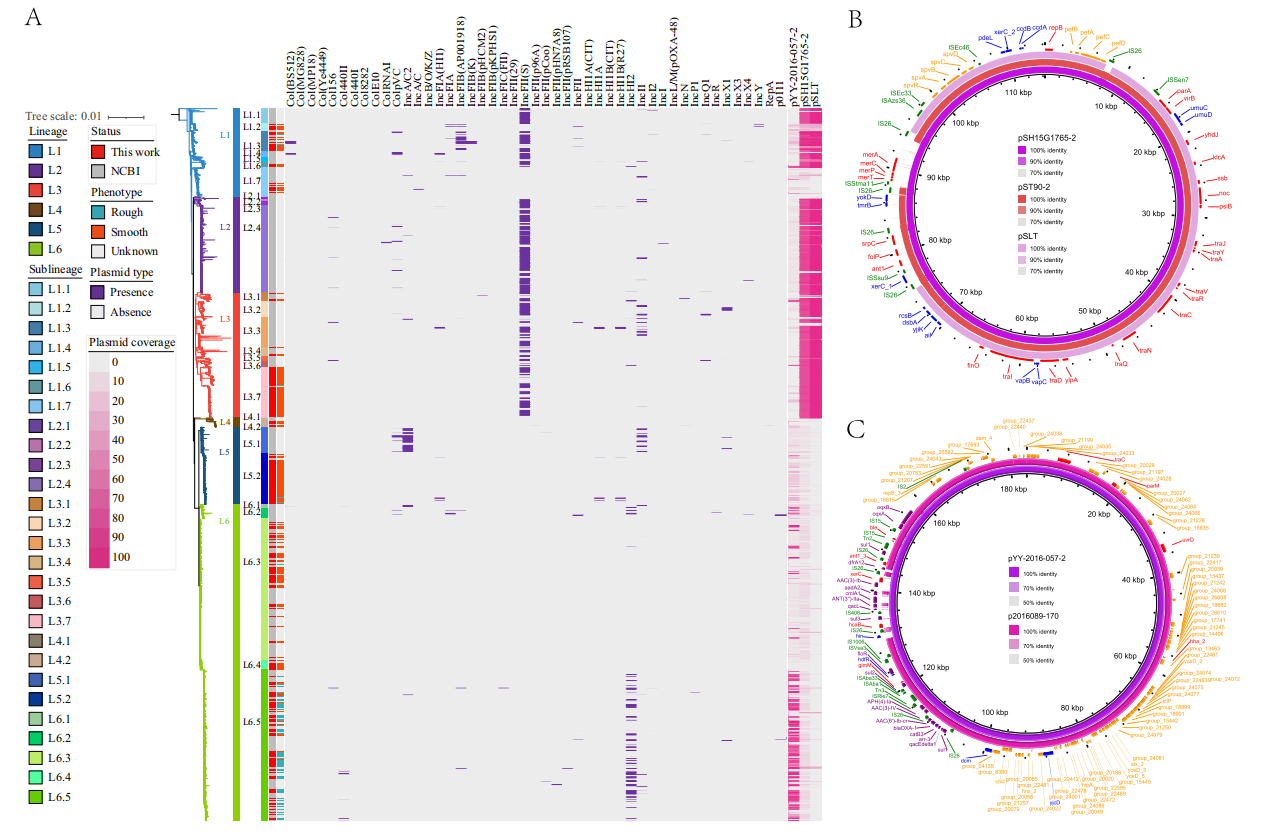
Figure 3: Overview and comparison of plasmids related to antimicrobial resistance and virulence factor genes
5 Pan-Genome and Whole-Genome Association Analysis
Pan-genome analysis identified 41,024 genes, and 87 genes significantly associated with rough colony isolates of sublineage L6.5 were selected. Genome-wide association studies (GWAS) identified 72 single nucleotide polymorphisms (SNPs) significantly associated with rough colony variants, 65 of which are located in protein-coding regions, and 7 are located in intergenic regions.
6 Multi-Omics Analysis
Genome and proteome sequencing were performed on both rough and smooth colony strains to analyze genetic and protein expression differences. The mRNA abundance of the csgBAC-csgDEFG operon was measured to validate the proteomic data, showing that the mRNA levels of the csgDEFG operon in the rough colony strain SF175 were significantly higher than in the control strain and smooth colony strains. To further determine the function of key genes and validate the association between genes and phenotypes, csgD knockout mutants (SF175-ΔcsgD,constructed by Ubigene) and mutants with a single nucleotide replacement in the csgD promoter (-44T > G) (SF175-csgDwith-44T > G) were constructed to observe the differences in colony morphology and biofilm formation ability between these mutants and the wild-type SF175 strain.
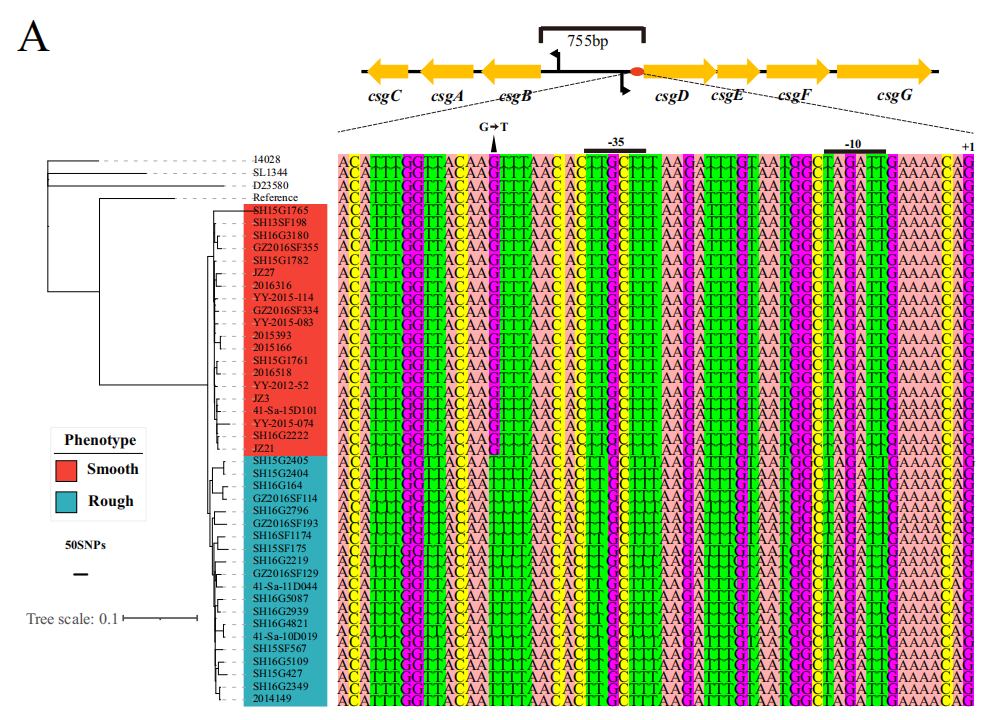
Figure 4: Phylogenetic tree of rough and smooth strains and nucleotide mutations in associated genes
Conclusion
This study provides a comprehensive and systematic investigation of a rough colony variant of S. Typhimurium. Through various analytical methods, it reveals the epidemiological distribution, biological characteristics, genomic structure, and genes related to antimicrobial resistance and virulence. The findings suggest that this variant is widely distributed in China and possesses multidrug resistance and strong biofilm formation ability. Its unique genomic features place it in a new phylogenetic lineage, which may pose a potential public health threat.findings provide important insights into the evolution and transmission mechanisms of S. Typhimurium and offer new strategies for controlling the spread of this pathogen.
References
Xiang, Ying et al. “Characterization of a Salmonella enterica serovar Typhimurium lineage with rough colony morphology and multidrug resistance.” Nature communications vol. 15,1 6123. 20 Jul. 2024, doi:10.1038/s41467-024-50331-y










