IF=14.3 | Ubigene KO Cells Assist in Revealing the Function and Mechanism of Targeting piRNA in Promoting Prostate Cancer Development

Prostate cancer (PCa) is the second most common cancer in males and the fifth leading cause of cancer death worldwide. Emerging evidence has demonstrated that genetic and epigenetic events, including genetic variants, non-coding RNAs (ncRNA),and N6-methyladenosine
(m6A) methylation, contribute to the development and progression of PCa.
Recently, a study by Meilin Wang and colleagues from Nanjing Medical University, published in Advanced Science (IF=14.3), titled “piRNA PROPER Suppresses DUSP1 Translation by Targeting N6-Methyladenosine-Mediated RNA Circularization to Promote Oncogenesis of Prostate Cancer”, identified a specific genetic variant, rs17201241, associated with increased expression of PROPER (a piRNA overexpressed in prostate cancer) in tumors and located within the gene, which increases the risk and malignant progression of PCa. In this study, SNORD48 gene knockout PC-3 cells constructed by Ubigene revealed the production mechanism of PROPER and the role of rs17201241.
Among various types of ncRNAs, PIWI-interacting RNA (piRNA) is a novel class of small ncRNA, initially found in germline cells with high abundance, demonstrating the most famous and critical regulatory function in silencing transposons and maintaining genome integrity during gametogenesis and fertility. However, some studies have also reported additional regulatory roles of piRNAs in cancers originating from different cell types, such as reproductive or somatic tissues. For example, piR-823, piR-001773, and piR-017184 have been shown to play critical roles in the epigenetic regulation of cancer progression. Previous studies have indicated that piRNAs are highly expressed in various tumors and exhibit numerous genetic variations, suggesting their potential role in carcinogenesis. However, the causal relationship and molecular mechanisms of piRNA-related SNPs in cancer risks, including PCa, remain largely unknown.
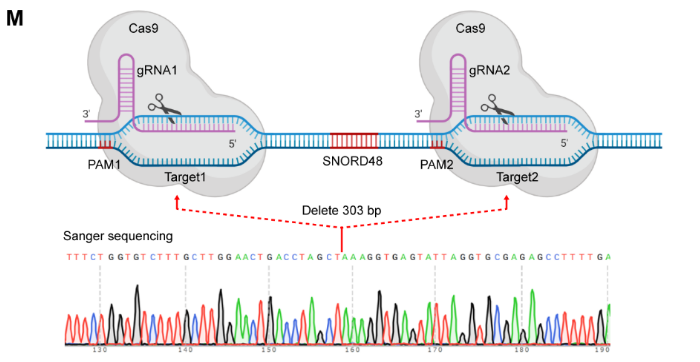
Figure 1 SNPRD48 knockout by CRISPR/Cas9
Furthermore, increasing transcriptomics evidence has highlighted the key role of m6A in malignancies. m6A modification is generally enriched around translation stop codons and 3'-untranslated regions (UTRs) to regulate mRNA. The dynamic m6A-modified sites are recognized and acted upon by various reader proteins, such as YTH domain-containing proteins and heterogeneous nuclear ribonucleoprotein (hnRNP) family proteins, which regulate mRNA splicing, export, stability, and translation. YTH domain family protein 2 (YTHDF2), a critical m6A reader, preferentially recognizes m6A and recruits RNA decay enzymes or bridging proteins, triggering the rapid degradation of mRNA with m6A modifications and interfering with translation. However, the stability and translation regulation mechanisms of m6A-modified mRNAs involving ncRNAs in PCa remain unclear.
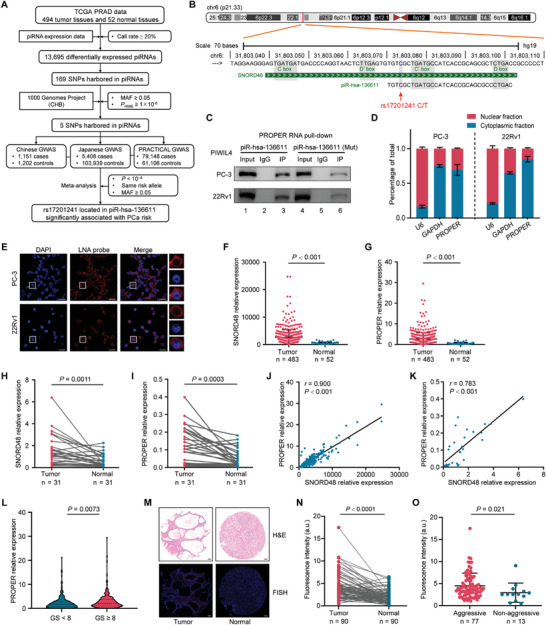
Figure2 Identification and expression analyses of PCa risk-associated piRNAs
Considering the complexity of post-transcriptional modifications in gene expression regulation, it is essential to integrate these modifications with classical regulatory layers, such as those involving well-explored pathways like the MAPK signaling pathway, to explain tumorigenesis mechanisms. Dual-specificity phosphatases (DUSPs) are key regulators in cell signaling pathways, especially the MAPK pathway. These enzymes dephosphorylate threonine and tyrosine residues on MAPKs, regulating their activity to ensure precise cellular responses to various stimuli. DUSP plays a crucial role in controlling cell proliferation, differentiation, and apoptosis, maintaining cellular homeostasis. By regulating MAPK signaling, DUSP1 can influence essential processes for metastasis, such as cell migration, invasion, and epithelial-mesenchymal transition (EMT). In the context of PCa, DUSP1 is downregulated, and its impact on tumor progression and metastasis is significant. However, the post-transcriptional modifications and regulation of DUSP1 itself remain poorly understood.
In this study, combined GWASs enabled us to find the PCa risk-associated variant rs17201241, residing in PROPER (piRNA overexpressed in prostate cancer), which is aberrantly highly ex-pressed in advanced PCa. We identified an unexpected RNA epigenetic regulatory mechanism by which PROPER is assembled with m6A binding protein YTHDF2 and YBX3 to form a piRNA-induced silencing complex (pi-RISC), thereby inhibiting translation and promoting degradation of DUSP1 with the translation initiation factor EIF2S3 in an mRNA-looping manner. Therefore, this study not only reports a previously unknown function of piRNA in post-transcriptional regulation but also providesnew therapeutic opportunities for targeting the piRNA-mediated translation machinery in cancer.
Reference:
Ben, Shuai et al. “piRNA PROPER Suppresses DUSP1 Translation by Targeting N6-Methyladenosine-Mediated RNA Circularization to Promote Oncogenesis of Prostate Cancer.” Advanced science (Weinheim, Baden-Wurttemberg, Germany) vol. 11,33 (2024): e2402954. doi:10.1002/advs.202402954