Published on: September 10, 2024
Views:
--
What is the multi-genotype of KO cells?

Many researchers may encounter a situation during the genotyping of knockout (KO) cells where the sequencing chromatogram of the target site of the single-cell clone shows overlapping peaks, or even more than two peaks. What is going on here? And can this clone still be used? Today, Ubigene will discuss the multi-genotype issue of KO cells.
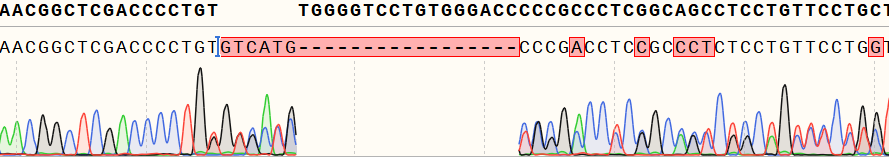
Figure 1 Genotype identification of the single-cell clone shows overlapping peaks
Q1: Why do different genotypes arise after editing?
Let's first review how the CRISPR/Cas9 technology works: The CRISPR/Cas9 system uses sgRNA to guide the Cas9 enzyme to a specific location in the genome, creating a double-strand break (DSB). Because cells cannot survive long with broken DNA, the process of repairing the break begins soon after the DNA is cut. Non-homologous end joining (NHEJ) is the main repair pathway for DSBs in most eukaryotes[1]. The NHEJ repair process is prone to errors, which can lead to random insertions or deletions of bases (indels). If indels of non-triplet length occur, they can cause frameshift mutations during gene translation, disrupting protein function and thereby achieving gene knockout.
Since NHEJ repair is random, the outcome of repair can differ for each allele. In actual gene editing operations, the process by which sgRNA and Cas9 enzyme bind and act on the genome is not completely uniform; it has a degree of randomness, possibly leading to differences in the DNA break location and repair results for the same allele[2]. Additionally, other intracellular molecules and environmental factors can interfere with the repair process, further increasing the possibility of genotype diversity.
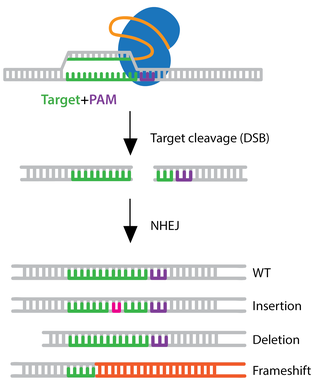
Figure 2 CRISPR/Cas9 non-homologous end joining (NHEJ)[3].
Q2: Why can there be more than two genotypes?
Mammalian normal somatic cells are diploid, but most cell lines used in experiments are cancer cells. During their development, cancer cells may undergo complex genomic instability and chromosomal abnormalities (such as chromosomal rearrangements, duplications, deletions, or additions), adapting better to various environmental conditions and gaining growth advantages. These abnormalities lead to a non-diploid state, often presenting as aneuploidy or polyploidy. Chromosomal abnormalities can lead to changes in the copy number of some genes, resulting in more than two alleles. The following image shows a karyotype of HeLa cells, where the number of chromosomes varies significantly, with chromosome 5 reaching as many as eight copies[4].
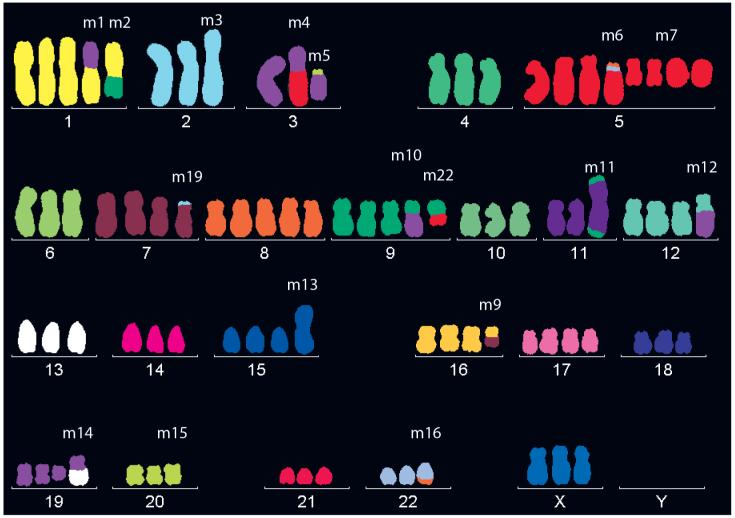
Figure 3 HeLa cells have more chromosomes than normal cells.
Gene editing tip: Before starting an experiment, you can find out the copy number of the target gene in the cell, which helps understand the genotype of KO cells and the difficulty of obtaining homozygotes. We recommend using the Gene Copy Number Assessment System on the Red Cotton website (https://www.rc-crispr.com/tools/gene_copy_number.html). This tool can calculate the average copy number of different genes in cell lines based on genomic sequencing results and karyotype data. The higher the copy number, the more difficult it is to obtain gene-edited homozygotes, which can be used for comprehensive project difficulty assessment.
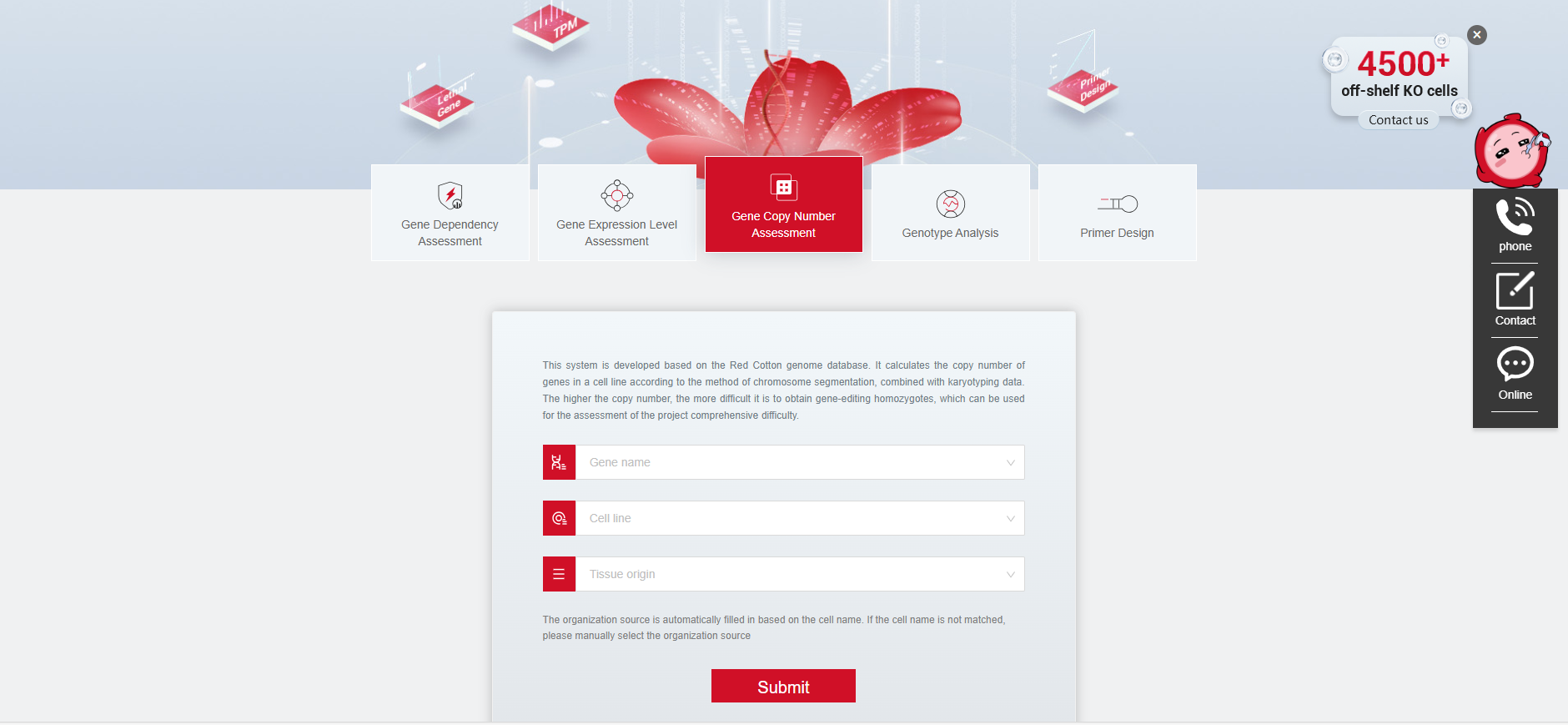
Figure 4 Ubigene’s Gene Copy Number Assessment System
Q3: How can different genotypes be distinguished?
Here are three commonly used methods:
1. Sanger Sequencing + Genotype Analysis Tool
Amplify the target region using PCR, and perform Sanger sequencing on the PCR product. Then, you can use another tool on the Red Cotton website: the Genotype Analysis System (https://www.rc-crispr.com/tools/gas.html) to analyze the sequencing results. This method is convenient and fast. You only need to sequence the PCR product, and with the help of the genotype analysis tool, you can quickly distinguish different genotypes and editing efficiency, greatly improving identification efficiency and reducing experimental costs.
Figure 5 Ubigene’s Genotype Analysis System
Case Study: Two genotypes of the diploid single-cell clone (both alleles are edited, but the deletions differ).
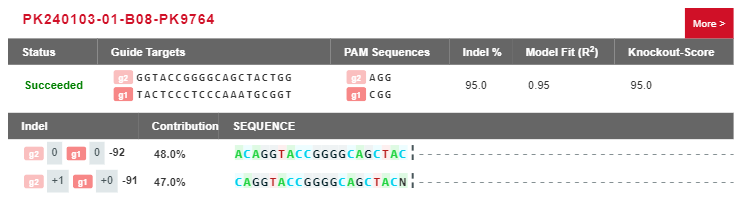
Figure 6 Result of Ubigene’s Genotype Analysis System
2.TA Cloning/Topo Cloning + Sanger Sequencing
Amplify the target region using PCR, and select the appropriate cloning method based on the PCR product's status. PCR products generally lack phosphorylation at the 5' end and can be directly used for Topo cloning or TA cloning after adding an "A" tail[5]. After cloning, screen clones and perform Sanger sequencing. This method distinguishes different genes by ligating PCR products to cloning vectors, but clone screening is required, making the process somewhat cumbersome.
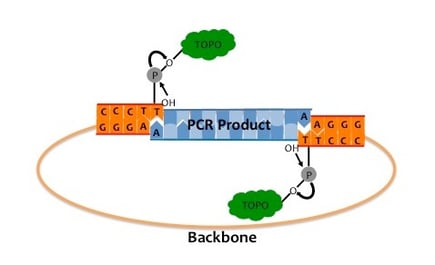
Figure 7 Topo cloning
Molecular Biology Knowledge
TA Cloning: Utilizes certain DNA polymerases (like Taq polymerase) to add adenine (A) non-template-dependently at the 3' end of PCR products, allowing the PCR product to be ligated to the thymine (T) at the 3' end of the linearized vector through complementary pairing[6].
TOPO Cloning: TOPO cloning is a one-step technique where a single enzyme (topoisomerase I) inserts DNA fragments into a linearized vector. TOPO cloning is suitable for blunt or sticky-ended PCR products and does not require the addition of an extra "A" tail.
3. NGS Sequencing
NGS sequencing can quantitatively assess genomic editing in the target sequence and also be used to check off-target effects. First, amplify the target region using PCR, purify and recover the PCR product, create an NGS library, and perform sequencing[7]. This method is relatively simple; you can give the PCR product to a sequencing company for subsequent steps, but it is more costly.
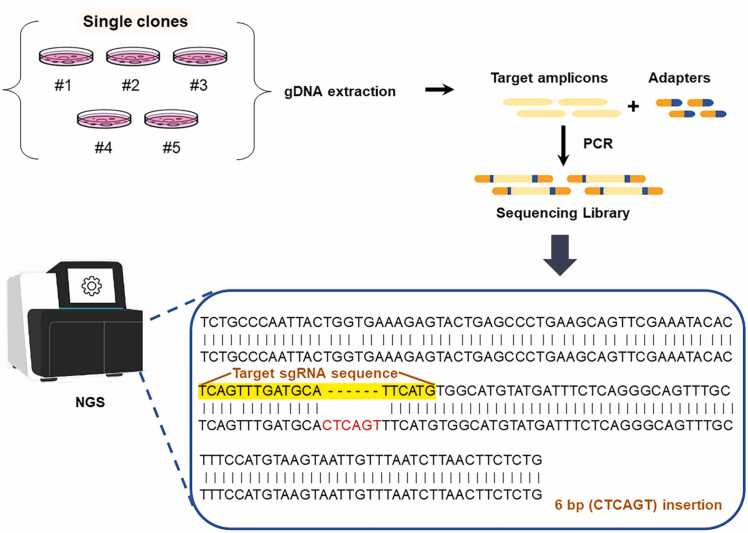
Figure 8 NGS sequencing on single-cell clones[7]
Q4:Can multi-genotype KO cells be used?
The main types of cell genotypes identified in gene knockout experiments include:
1)Wild Type (WT): All alleles of the target gene remain unedited and are consistent with wild-type cell lines. These cells can be used as negative controls in experiments.
2)KO Heterozygote (HE): Some alleles of the target gene are WT, and some are KO frameshift genotypes. These cells can be used as knockdown cells (similar to the interference effect of RNAi), especially when complete gene knockout is lethal. If heterozygous KO has a phenotype, it can be directly used in publications.
3)KO Homozygote (HO): All alleles of the target gene are KO frameshift genotypes, and the genotype is completely consistent. This is the most ideal genotype!
4)Compound Heterozygote: All alleles of the target gene are KO frameshift genotypes, but the deletion differs among alleles, such as one allele lacking 1 bp and another lacking 5 bp. Since each allele undergoes frameshift mutation and achieves KO effect, it can be equivalent to the third genotype and broadly called "KO homozygote," which can be normally delivered as KO clones.
That's all for today. After reading this, do you feel more familiar with the phenomenon of multi-genotype KO cells? If you have more questions related to gene-edited cells, feel free to contact us, we will arrange the frequently asked questions right away!
References:
[1]Chang, Howard H Y et al. “Non-homologous DNA end joining and alternative pathways to double-strand break repair.” Nature reviews. Molecular cell biology vol. 18,8 (2017): 495-506. doi:10.1038/nrm.2017.48
[2]https://blog.addgene.org/crispr-101-validating-your-genome-edit
[3]https://www.addgene.org/guides/crispr/#design-grna
[4]Smirnov, E et al. “NORs and their transcription competence during the cell cycle.” Folia biologica vol. 52,3 (2006): 59-70.
[5]Giuliano, Christopher J et al. “Generating Single Cell-Derived Knockout Clones in Mammalian Cells with CRISPR/Cas9.” Current protocols in molecular biology vol. 128,1 (2019): e100. doi:10.1002/cpmb.100
[6] https://www.takarabiomed.com.cn/QuestionAnswer.aspx
[7]Hong, Taeyeon et al. “Guide for generating single-cell-derived knockout clones in mammalian cell lines using the CRISPR/Cas9 system.” Molecules and cells vol. 47,7 (2024): 100087. doi:10.1016/j.mocell.2024.100087















