How to design gRNA and gRNA expression vector

The expression vector can drive high-level constitutive transcription of a user-selected gRNA sequence to achieve highly efficient CRISPR targeting when used in conjunction with Cas9 nuclease. A routine gRNA expression vector system would be simple and faster, cost-effective, efficient, and highly customizable. The development of a high-quality gRNA expression vector has been revolutionary and simplify CRISPR experiments.
Principle of gRNA design:
Reporters can be fluorescent and luminescent proteins such as green fluorescent protein (GFP) that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range, and the enzyme luciferase, which catalyzes a reaction with luciferin to produce light, in other cases, it can also be a tag that is to fused with the interested gene, such as glutathione S-transferase (GST), histidine (HIS), and flag tag, which will allow antibody-based detection and affinity-based isolation of the interested gene products.
CRISPR consists of an endonuclease protein (Cas9) for targeting a specific sequence RNA called guide RNA (gRNA) for guide endonuclease proteins to the target region. In CRISPR mediated genome editing, one of the key issue is to design and select the gRNA. Before starting to design and select gRNA, few questions need to address when choosing a gRNA: First, Does a given gRNA sequence exactly match your genomic target? If not the variation between a given gRNA sequence and genomic target may reduce the gRNA activity and increases the off-target effect. However, Cas9 enzyme can tolerate up to four mismatches (Between the gRNA and its genomic target), and one nucleotide gap. Thus, in a tolerable mismatches situation, cleavage can occur in the target site as well as a significant portion of off-target activity also occurs within gene coding sequences. Therefore, the off-target activity can lead to inaccurate interpretation of the CRISPR results and high off-target activity can cause depletion of the Cas9 enzyme or even cell death. Second, which species or variant of Cas9 (S. pyogenes, S. aureus, etc.) was this gRNA sequence designed for? A given gRNA sequence may only be compatible with a single species or PAM binding variant of Cas9. For instance, wild-type SpCas9 must be used with targets that are upstream of a 5' NGG 3' PAM sequence. Third, Which CRISPR application is this gRNA sequence compatible with? CRISPR knockout experiments use targeting sequences within exons, whereas CRISPR activation or repression experiments use targets within promoters.
There are multiple online tools are available to design guide RNAs with high editing efficiencies and low off-target effects. When designing gRNA by using online Bioinformatics tools, the following information is considered to choose the best gRNA. The gRNA with the lowest levels of Off-target, Optimum GC count, PAM (Protospacer Adjacent Motif), efficiency, different transcription variants of a gene, gRNA position, and no stable gRNA stem-loop. We’ve profiled the top CRISPR design tools that will simplify your experiments.
1. DESKGEN Cloud is the most comprehensive gRNA design software available, doing everything the MIT tool can and much more. By aggregating scoring functions from academic literature and other CRISPR tools, DESKGEN keeps you up to date with best practices in the field.
2. Benchling is designed with real scientist workflows in mind, from the most cutting-edge techniques to the most reliable everyday ones. Benchling product supports automated Gibson, Type IIS, and Digestion and Ligation cloning, so scientists do not need to switch between multiple tools.
3. MIT CRISPR design tool is a web tool crafted to simplify the process of CRISPR guide selection in an input DNA sequence by (i) discovering possible off-targets genome-wide, (ii) highlighting guides with high target specificity, and (iii) flagging guides with numerous or genic off-targets in target genomes.
4. CRISPR Gene Editing Designer (Red CottonTM) a computational tool to assist researchers in performing CRISPR-Cas9 experiment optimally. Red CottonTM provide maximize on-target activity (guide efficiency) as well as minimizing potential off-target effects (guide specificity) by analyzing the features of the target site. It is a very simple and user-friendly designing tools. The only species name, gene name/ID, and cell name are required to operate the system. The researcher can get valuable information about genes and cells within one minute with 3 knockout strategies, analysis of gene transcripts and sequence complexity, more than 800 cell line parameters, and 3000 successful knockout information.
Design gRNA with Red Cotton™ tools
1. Design gRNAs using online tools Red CottonTM (https://www.rc-crispr.com/.).
2. Use the species name, gene name/ ID, and cell line name to generate potential gRNA sequences.
Note: All the potential gRNAs against the gene are created. The quality of the potential gRNAs will be reported as Score indicated on the on-target cutting activity.
3. Consider the Off-target activity in the gRNA selection when ranking the gRNAs.
Note: Optimal guides are those with the greatest on-target efficiency and the least off-target activity. To disable a gene using Cas9 cutting, gRNAs are designed to target the first 10 - 50% of the coding sequence downstream of the initiating codon (as gRNAs targeting the 3’ end of the gene are less effective).
4. Ensure no internal BsmBI sites are present if oligonucleotides are being used for the Golden Gate cloning, as this will result in the annealed oligonucleotides being digested.
Using the Red CottonTM web tool (https://www.rc-crispr.com/), find candidate gRNA target sequences in the genomic region of interest. The output window shows 23 bp genomic sites of the form 5'-N20NGG-3' within your target region. These sites may reside on the + or - strand.

After select and synthesize gRNA oligos, it is necessary to construct a gRNA expression vector. The expression vector will be used for the transfection of cells.
gRNA expression vector (Plasmid):
The plasmid gRNA expression vector is a highly efficient tool for transfection-based delivery of target site-specific gRNA sequences into mammalian cells. Delivering plasmid vectors into mammalian cells by transfection is one of the most widely used procedures in bio-medical research. The plasmid transfection approach remains the workhorse of gene delivery due to its technical simplicity as well as good efficiency in a wide range of cell types. A key feature of transfection with plasmid vectors is that it is transient, with only a very low fraction of cells stably integrating the plasmid in the genome (typically less than 1%).
To achieve CRISPR-mediated gene targeting it is essential for the target cells to co-express both Cas9 as well as the target site-specific gRNA at the same time. One strategy of transfection is the vector carrying both Cas9 and the gRNA sequence and transfect the target cells, this transfection system is termed as an all-in-one vector transfection system. The vector carries reporter and selection markers (eg. EGFP and Puro).
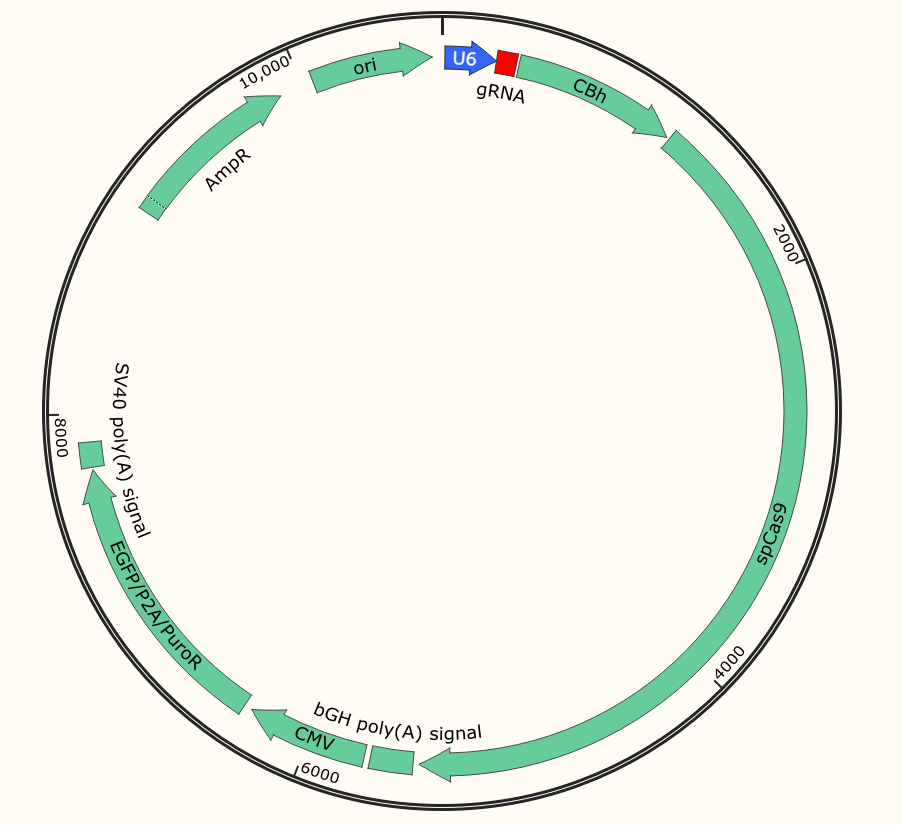
Fig 1: All in one plasmid vector
The other strategy is plasmid-based transfection by using separate vectors for driving Cas9 and gRNA expression (Cas9 only and gRNA only vectors respectively).
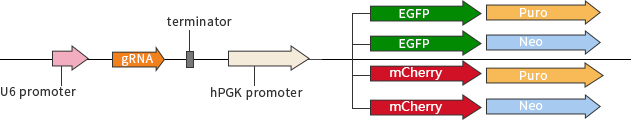
Fig 2: plasmid vector with single gRNA
There are several advantages of using separate vector systems. Separate vectors offer the flexibility of combinatorial usage of different gRNA expression vectors in conjunction with a variety of Cas9 variants (wild type nuclease, nickase, nuclease-dead) depending upon the user’s experimental goal. Using a separate gRNA vector allows cells or organisms stably expressing high levels of Cas9 to be transfected with different gRNA sequences targeting either the same gene or different genes. This provides the opportunity for comparing the efficiencies of different gRNA sequences in parallel at CRISPR-mediated gene targeting in cells or organisms with comparable and high levels of Cas9 expression.
Application:
1. The vector carrying reporter marker (eg. EGFP, Mcherry) can be used to monitor transfection efficiency and/or to enrich/isolate transfected cells by flow cytometry.
2. Selection markers (eg. Puro, Neo) confer resistance to antibiotics and helps for artificial selection.
Since the release of Red Cotton™ CRISPR Gene-editing Design System, it has brought the benefits to researchers from more than 20 countries and regions and assisted their gene study researches. In 2021, Ubigene continues to achieve our goal – “Make genome editing easier” and to bring more benefits to you.
Ubigene has set-up our gRNA plasmid bank!
10000 in-stock plasmids available! Only $80.
Deliver in one 3-5 week days.
Most economical and lowest cost for gene KO.
Check out your interested genes from our database now.
Click Here >
gRNA plasmid bank plus Red Cotton system, the best and easiest kit for gene KO.
Over 600 types of gene plasmids are now ready for you!
Sign-up Red Cotton™ now and get your free vector. The first 50 registers can get the FREE plasmid each week.
Click the link to check out our free plasmids
Click Here >









