Guideline of reporter cell line. How to pick a reporter?
KI or overexpression? All in Ubigene
Reporter gene refers to genes encoding proteins that are easy to be detected (such as luminescence, dyeing, antibiotic resistance, etc.), and the phenotypes generated by these genes do not exist in the original receptor cells, tissues, organs, or individuals. Common reporter genes include fluorescent protein (EGFP, mCherry, etc.), luciferase (Fluc, Rluc, etc.) β-Galactosidase gene (LacZ), etc.
LacZ can be dyed blue by X-gal, which is often used for gene expression research of whole embryos or tissue slices, but less used for cell experiments. Luciferase needs to add luciferin substrate to produce luminance, while fluorescent protein can spontaneously fluoresce under the irradiation of specific wavelengths of excitation light. These two kinds of reporter genes are commonly used in cell transfection experiments, gene expression detection, promoter/enhancer research, in vivo imaging, protein localization, cell tracing, etc. In this article, Ubigene introduces the common applications and selection strategies of these two kinds of reporter genes at the cell level, to provide some ideas for your research design.
Applications:
1.Using luciferase or EGFP stable cell line to construct a mouse model of transplanted tumor.
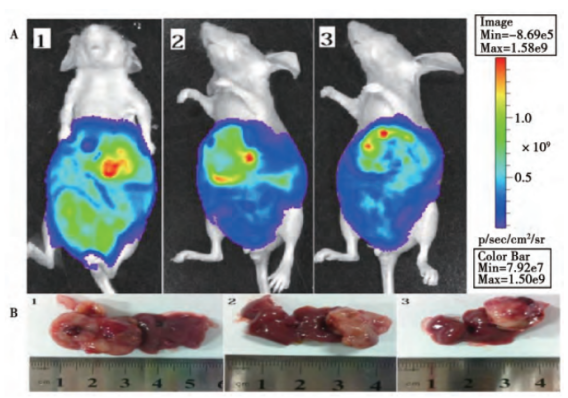
To construct cell lines that can express luciferase or fluorescent protein, and transplant them into animals. And quantitative, qualitative and in vivo tracking can be carried out through molecular imaging technology or scintillation counters. Reporter cell line is commonly used in cancer and tumor cell transplantation study. Xiangming Zhai et al. [1] constructed a lentivirus vector PTYF-CMV-Luc-2A-RFP-PGK containing luciferase and fluorescent protein, and then infected the liver cancer cell line SMMC7721 after lentivirus packaging. They screened it with Puromycin, and obtained a stable cell line by the limited dilution method. Then, Luc-2A-RFP cells were injected into the liver tissue of nude mice to establish an animal model of in situ liver cancer. The distribution of liver cancer cells in living animal was successfully observed by the in vivo fluorescence imaging system, laying a foundation for the treatment of liver cancer (Figure 1, Figure 2).
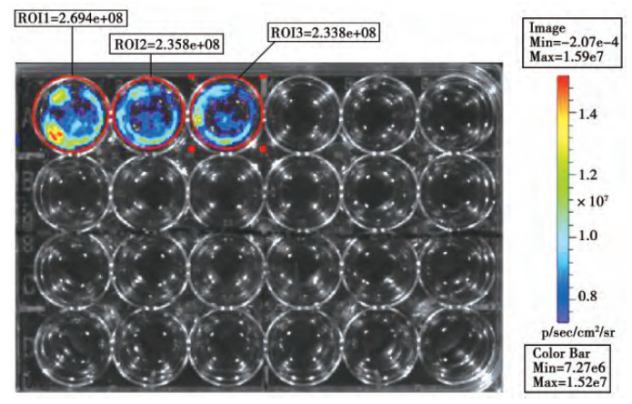
Figure 1. In vitro biofluorescence detection
Figure 2. In vivo biofluorescence detection and corresponding liver tumor volume detection
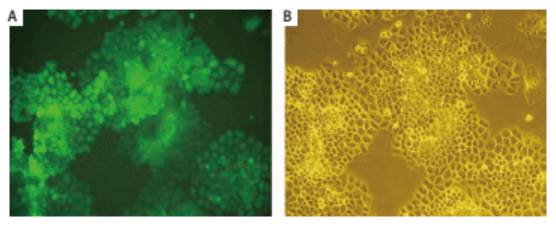
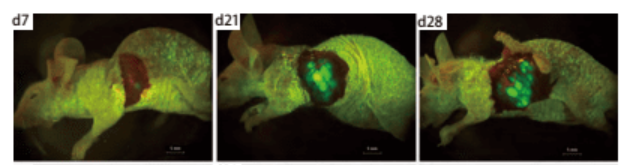
Shuzhen Wei et al. [2] introduced EGFP gene into human lung cancer cell line NCI-H460 by retroviral transduction, constructed a lung cancer cell line that can stably and highly express EGFP, and carried out subcutaneous tumorigenesis in nude mice. Then, the orthotopic transplantation model of mouse lung cancer was established by orthotopic transplantation, and tumor growth was observed by using small animal living fluorescence imaging system. This model can be used for tumor drug screening and other related studies (Figure 3, Figure 4).
Figure 3. Monoclonal H460 cells stably expressing EGFP
Figure 4. In vivo fluorescence imaging of orthotopic transplanted tumor in nude mice
Ubigene provides over a hundred kinds of Luciferase and EGFP stable cell lines with expression data. Only $1590 for our custom stable cell line service, speed up your experiment process! More than 200 kinds of cell lines including THP-1/RAW264.7/Hep G2 and other challenging cell lines are available. Stable expression and high homogeneity. Contact us for more details of our service>>
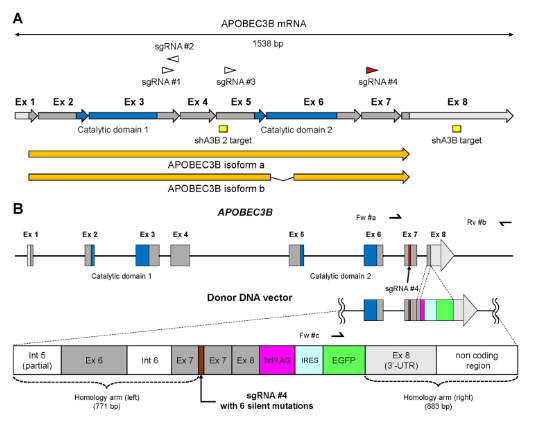
2.Construct report cell lines for drug screening by CRISPR/Cas9 mediated in situ knockin of EGFPAPOBEC3B (A3B) is a DNA editing enzyme that induces genomic DNA mutations in multiple myeloma and various cancers. In order to facilitate the study of the function of A3B, Yamazaki et al. [3] knocked 3xFlag-IRES-EGFP into the 3 'end of A3B through CRISPR/Cas9 technology, and constructed three kinds of A3B reporter myeloma cell lines (U266, RPMI8226, AMO1). Through knockdown verification of shRNA lentivirus targeting A3B gene, it was found that when A3B mRNA was down-regulated, EGFP fluorescence intensity also decreased, which confirmed the feasibility of reporting cell model and could be used for subsequent anti-cancer drug testing (Figure 5).
Figure 5. Targeting map of EGFP Knockin A3B gene
Ubigene provides knockin cell line construction service to knockin tags, luciferase, and fluorescent proteins, and to facilitate drug screening, disease model construction, promoter expression activity research, etc. Reporter gene KI in more than 200 kinds of cell lines only takes $9480.
Visit us for more service details>>
3.Detection of promoter expression activity with Luciferase or EGFP
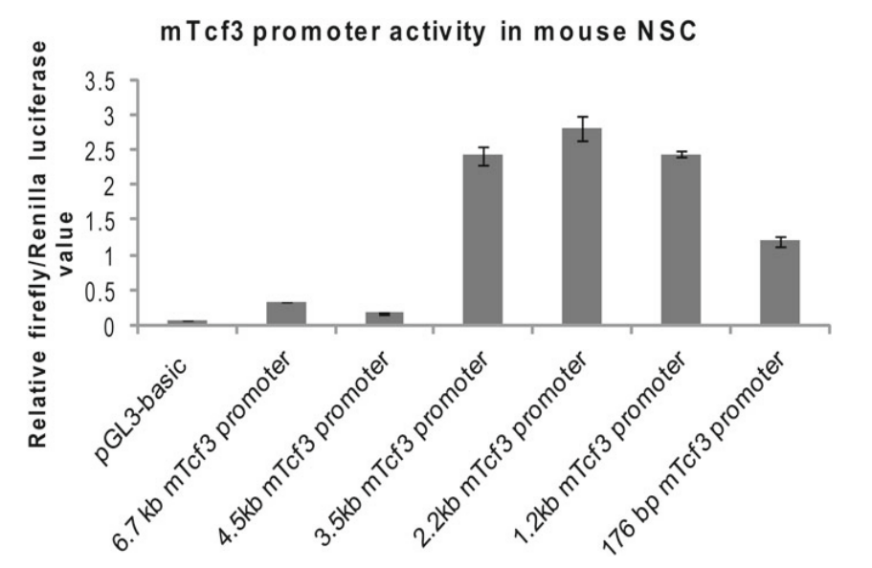
Compared with traditional methods such as Western Blot and ELISA, reporter genes can monitor the expression level of target genes or promoter activity in vivo. Generally, the method of inserting fluorescent protein or luciferase after the promoter sequence is used for monitoring promoter activity. For example, Nina Solberg and Stefan Krauss [4] cloned the mTcf3 promoter fragment into the pGL3-basic report vector containing Fluc cDNA, and studied the activity of the mTcf3 promoter DNA fragment cloned in mouse embryonic-derived neural stem cells (NSCs) by comparing the activity of firefly luciferase with that of the Ranilla luciferase (reference). It was found that there were regulatory elements inhibiting promoter activity in the truncated fragment between 4.5 kb and 3.5 kb promoter fragments (Figure. 6).
Figure 6. Activity analysis of mTcf3 promoters with different lengths
4.Study on the interaction between miRNA and 3'UTR by dual-luciferase reporter systemAt present, the conventional idea for studying miRNA target genes is to predict miRNA target genes using bioinformatics methods such as gene prediction database, and then verify them through the dual-luciferase reporting system. In the step of dual-luciferase reporting system validation, it needs to connect the 3'UTR sequence of the target gene to the 3 'end of luciferase on the vector, and then determine the target gene that acts with miRNA by comparing the changes in the luminous intensity before and after miRNA overexpression. For example, through expression array analysis, Marcos et al. [5] assumed GSK3B as the target gene of miR-548q and miR-1185-1 which are related to weight loss intervention, and verified it with a dual-luciferase reporting system. They cloned the GSK3B specific 3'-UTR binding region downstream of the firefly luciferase gene, constructed a vector and co-transfected it with miR-548q and miR-1185-1 simulants into HEK-293T cells, verifying that GSK3B is the target gene of miR-1185-1 (Figure 7).
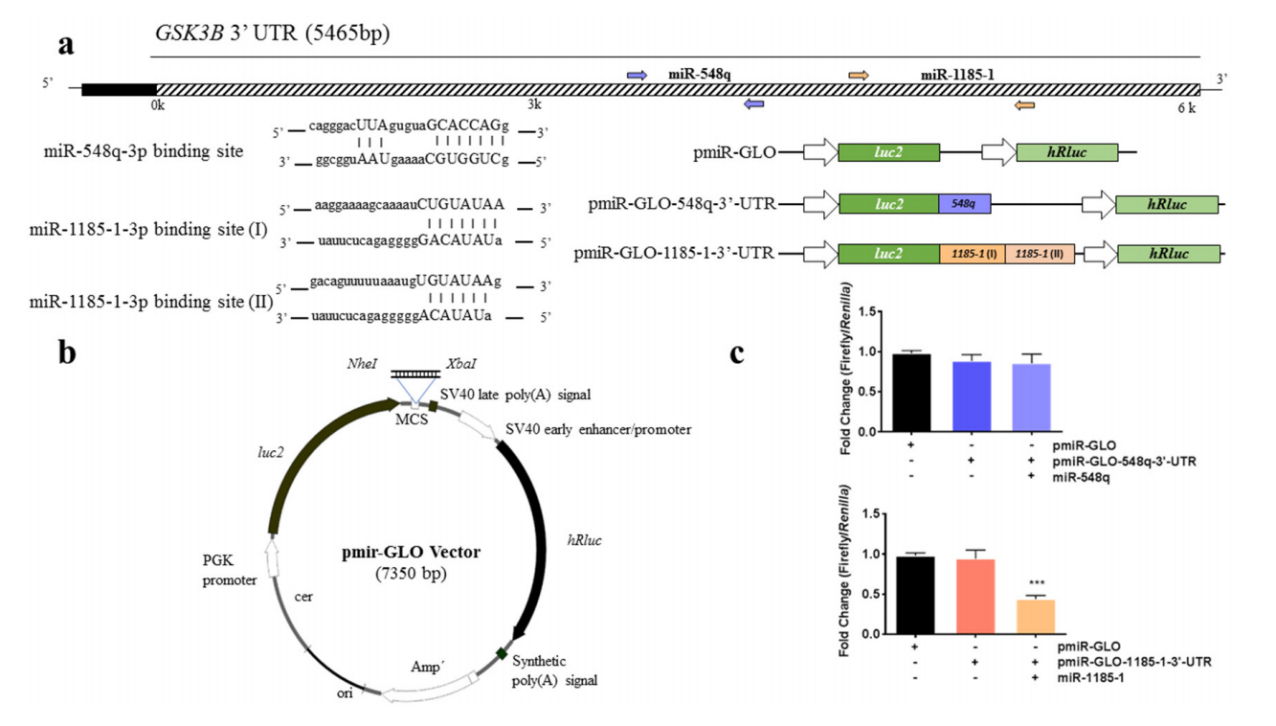
Figure 7. Binding analysis of GCK3B and miR-548q and miR-1185-1
In addition, reporter genes also have applications in protein interaction, RNA interference research, positive clone screening, etc. In the above cases, we found that the reporter cell line can be divided into two types from the expression genes, one is luciferase reporter gene, and the other is fluorescent protein reporter gene. There are also two construction methods of reporter cell lines, one is stable transfection, the other is in situ knockin (KI). What are the differences between these reporter genes and construction methods, and how to choose the right one for your research? We have sorted out the following key points for you:
Compared with fluorescent protein, luciferase has the advantages of high sensitivity, low background interference, high fluorescence intensity, strong penetrability, and easy quantification. It is more often used for promoter activity detection, miRNA function research, mouse tumor transplantation model construction, etc. The advantage of fluorescent protein is that it is convenient to observe without adding substrate, has low toxicity, multiple colors are available for selection, and the image is beautiful. It is suitable for polygene reporting systems, and is often used as a marker for stable cell lines, endogenous gene tracing markers, protein localization markers, and in vivo imaging. In general, both of these two kinds of reporter genes can be used in most cases. Many researchers also use the method of combining luciferase and fluorescent protein to co-express and construct the multi-functional report cell line. As for the construction method, in general, endogenous gene tracing and protein localization need to be constructed in situ by KI, while for other applications, stable overexpression can meet the experimental requirements.\
Related products
Over a hundred Cas9 stable cell lines>> stably express Cas9 protein, and transfected with gRNA and donor DNA can achieve gene knockin!
Reference
[1]翟向明, et al."携带荧光素酶基因肝癌细胞株的构建及在肝癌原位移植瘤模型中应用." 生物技术 28.01(2018):71-76. doi:10.16519/j.cnki.1004-311x.2018.01.0013.
[2]魏淑珍,etal."EGFP标记的人肺癌裸鼠原位移植模型的建立." 中国肺癌杂志 13.07(2010):670-675.
[3]Yamazaki H, Shirakawa K, Matsumoto T, et al. APOBEC3B reporter myeloma cell lines identify DNA damage response pathways leading to APOBEC3B expression[J]. PloS one, 2020, 15(1): e0223463.
[4]Solberg N, Krauss S. Luciferase assay to study the activity of a cloned promoter DNA fragment. Methods Mol Biol. 2013;977:65-78. doi: 10.1007/978-1-62703-284-1_6. PMID: 23436354.
[5]Garcia-Lacarte M, Mansego ML, Zulet MA, Martinez JA, Milagro FI. miR-1185-1 and miR-548q Are Biomarkers of Response to Weight Loss and Regulate the Expression of GSK3B. Cells. 2019 Nov 30;8(12):1548. doi: 10.3390/cells8121548. PMID: 31801236; PMCID: PMC6953011.