Reporter KI cell model application

Since it was developed in 2012, this gene-editing tool has revolutionized biology research, making it easier to study disease and faster to discover drugs. The CRISPR/Cas9 system has become synonymous with genome editing in today’s world due to its simplistic mechanism. The CRISPR system is a precise genome editing technique, which can create gene knockout or knock-in genome manipulations through the substitution of a target genetic sequence with a desired donor sequence. Reporter KI is one of the most common applications of CRISPR/Cas9.
What is reporter cell line and reporter gene?
Reporter cell lines are stable cell lines that have been labeled with reporters, allowing the visualization, tracking, and isolation of interested genes. A reporter cell line can be generated through exogenous expression of a cassette with an easy-to-check tag (e.g. GFP) driven by the promoter of the interested gene. However, due to the exogenous expression level can lead to artifacts, a reporter with endogenous expression is preferred. In this regard, reporter cell line generated by knock-in a tag (e.g. GFP) downstream of the promoter of the interested gene is optimal.
Reporters can be fluorescent and luminescent proteins such as green fluorescent protein (GFP) that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range, and the enzyme luciferase, which catalyzes a reaction with luciferin to produce light, in other cases, it can also be a tag that is to fused with the interested gene, such as glutathione S-transferase (GST), histidine (HIS), and flag tag, which will allow antibody-based detection and affinity-based isolation of the interested gene products.
Strategy for reporter cell line generation by knock-in
For the introduction of foreign DNA, the reporter gene are needed to be brought into the genome. Researchers can introduce a reporter coding sequence (e.g., eGFP) downstream of the promoter of the targeted gene (Fig 1B). Another strategy, the reporter coding sequence used to replace specific locus of the target gene and generate a truncated protein (Fig 1C). A reporter gene can be inserted into a genomic site immediately downstream of the coding region of an endogenous gene, and co-expressed bi-cistronically with the endogenous gene as a single transcript under the control of the native transcriptional regulatory machinery (Fig. 1D). Furthermore, reporter gene coding sequence can be introduced in N/C terminal before the stop codon and under endogenous promoter control (Fig 1E). The strategies are commonly used for generation of a knock-in cellular models are shown in Fig 1.
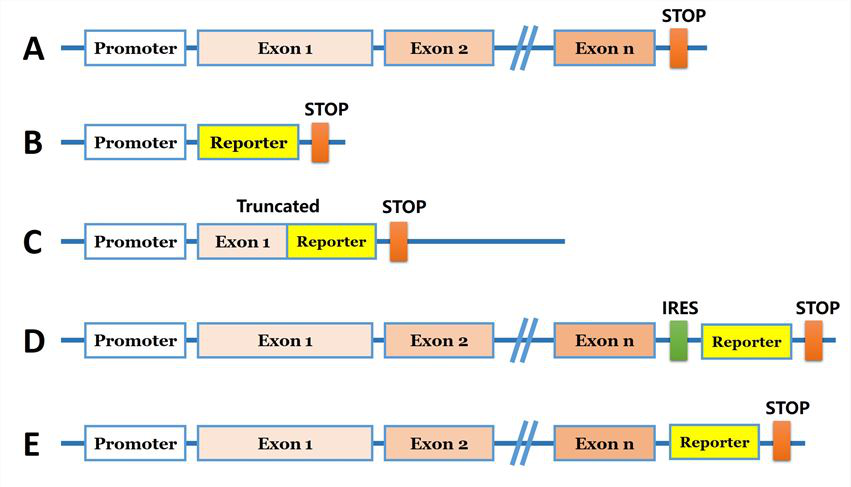
Fig 1: Strategies for reporter gene knock-in
Applications of reporter cell models
1. Transcriptional activity of gene promoter
Cell lines with endogenously tagged genes enable researchers to track protein production and localization in real time in live cells. These cell lines are also suitable for western blotting, protein pull down, affinity chromatography, immunocytochemistry, and flow cytometry among others. These can also be a good
solution when working with a protein for which there is no good antibody. The targeted gene coding sequence is replaced with a reporter gene coding sequence (e.g., EGFP), where the endogenous gene is knocked out as the reporter gene is simultaneously knocked-in via homologous recombination. Upon recombination, the promoter of the targeted gene will drive expression of the inserted reporter gene. An example, luciferase knock-in to study transcriptional regulation of the human SREBP1 gene. In this study, the researcher integration of a single copy of the exogenous luciferase gene into one allele of the genome and a 14 bp deletion of the targeted sequence in the other. Luciferase activity was directly correlated with the promoter activity of the endogenous SREBP1 gene in the HEK293-SREBP1- T2A-luciferase-KI cell line cell line.
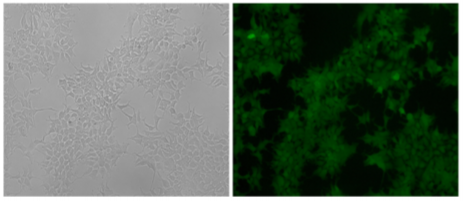
Fig 2: The HEK293 cell line expressed the exogenous eGFP reporter gene with undetectable expression of mCherry fluorescence after positive and negative selections with G418 and GCV.
2. Expression, localization, and transportation of proteins in cells
The visualization of a gene's expression can be realized by adding a tag sequence into the end (or the beginning) of the coding sequence of the targeted gene which will result in a fusion protein. As a result, expression levels of both the endogenous and recombinant proteins can be continuously monitored using fluorescence. The production of any protein of interest can be quantified based on the fluorescent reporter output of a cells. An example of knock-in, GFP-LC3 reporter in 293FT cells, where GFP is added to the N-terminal of endogenous LC3. Thus, the construction of the GFP-LC3 reporter knock in allowing GFP-LC3 fusion protein expression which is driven by the endogenous promoter without altering expression of the target gene. The endogenous GFP-LC3 reporter system showed accurate identification and quantification of cellular autophagy activity. The endogenous GFP-LC3 reporter system were verified by stress induced autophagy as well as drug (Rapamycin) induced/inhibit autophagy studies. As shown by the authors, free GFP protein is increase and its signal is still dispersed but stronger upon induction of autophagy, whereas GFP-LC3 become puncta like upon autophagy inhibition.
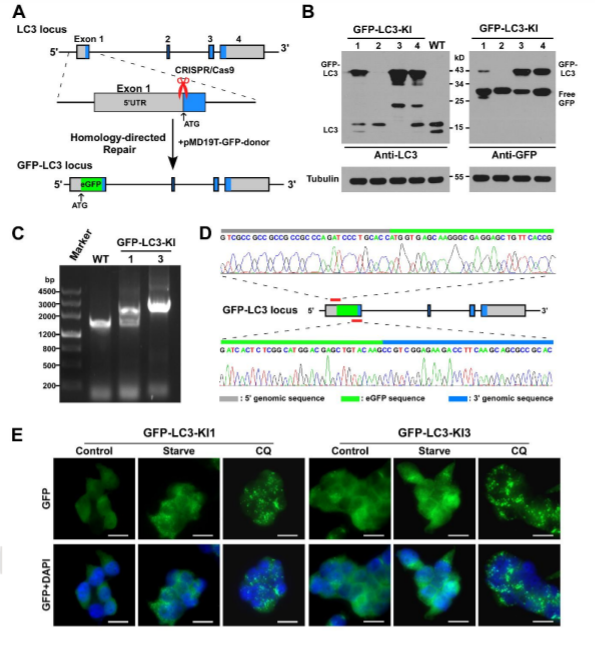
Fig 3: Generation of GFP-LC3 knock-in at the MAP1LC3B locus in 293FT cell.
3. Target identification and evaluation of drug candidates
Genetically engineered live cells lines containing reporters for the expression of single specific genes is useful for drug target identification and candidate evaluation. Thus, once a particular gene has been chosen as a target, the first step of the process is to clone the regions of DNA that regulate expression of that gene and fused to a highly sensitive reporter gene like the firefly luciferase gene, which generates a readily measurable light signal in response to changes in transcription of the target gene. When an engineered cell line treated with appropriate compound generate that generate a light signal which reflects a specific change in gene expression. Therefore, a reporter cell-based assays can allow the rapid screening of a large number of samples and excluded those compounds that either cytotoxic or fail to interact with the cell. An example of reporter cell line drug target evaluation, a HELA cell line was tagged with a nuclear protein (Histone 1-mtagBFP2), a cytoskeleton marker (β tubulin-mClover3), and a known autophagy receptor protein (SQSTM1-mRuby3) to screen for kinase inhibitors (with known targets) that induce the accumulation of cytoplasmic autophagic vesicles for evaluating the accumulation of autophagic vesicles. The cell was treated with two kinase inhibitors PLK1 inhibitor BI-6727 and PIM kinases inhibitor CX-6258 that increase the number of autophagic vesicles like hydroxychloroquine, a known autophagy inhibitor (Fig 4).
Fig 4: Validation of kinase inhibitors that induce the accumulation of autophagic vesicles in a triple labeled HELA cell line.
In summary, reporter cellular models generated by knock-in are robust for functional studies such as monitoring gene expression and subcellular localization and are useful for high-through-put drug screening.
Ubigene has modified over 100 types of cell lines with our CRISPR-U system. It has optimized the gRNA designing and targeting constructs backbone. The efficiency of splicing and recombining would be much higher than the traditional CRISPR. Now we offer an affordable reporter KI cell line service, only 9980 USD, positive clone deliverable!
Reference:
Establishment of a HEK293 cell line by CRISPR/Cas9- mediated luciferase knock-in to study transcriptional regulation of the human SREBP1 gene. Biotechnol Lett, 2018, (11-12):1495-1506.
CRISPR/Cas9 mediated GFP knock-in at the MAP1LC3B locus in 293FT cells is better for bona fide monitoring cellular autophagy. Biotechnol J, 2018, 13(11): e1700674.
A Versatile Vector System for the Fast Generation of Knock-in Cell Lines with CRISPR, 2020, doi: https://doi.org/10.1101/2020.02.06.927384










