[Research highlight] Ubigene
provides CRISPR lentivirus to facilitate Non-invasive diagnosis of BLCA
T1DMR and T2DMR DNA methylation
detection method
Background
Bladder cancer (BLCA) is one of the
most common cancers of the genitourinary system and the tenth most common cancer in the world. There are about
549,000 new cases and about 200,000 deaths every year. Among
them, 75% of patients were diagnosed as non-muscle invasive
bladder cancer (NMIBC) and the remaining 25% were muscle invasive bladder cancer (MIBC). The treatment methods
of these two kinds of tumors are completely different. Therefore, it is important to accurately classify BLCA
before determining the treatment strategy. Moreover, the choice of clinical treatment
for BLCA also depends on the accurate determination of tumor stage. Inaccurate stage will introduce unnecessary
surgical treatment and reduce the survival rate after treatment, and may also permanently reduce the quality of
life of patients. However, the traditional non-invasive diagnosis method for BLCA has low sensitivity and specificity, which makes it difficult to detect early
tumors, micro tumors, tumor residual foci and recurrent tumors, and to accurately distinguish between high-grade
(HG) and low-grade (LG) tumors, MIBC and NMIBC.
Therefore, Wang et al. from Zhongnan Hospital of Wuhan University studied the comprehensive characterization of DNA
methylation (DNAm) in BLCA. They found that DNA methylation is a specific marker of pan
cancer, and the detection of special DNA methylation can be used for cancer diagnosis and accurate classification of
subtypes. Therefore, the whole genome targeted sequencing of different tumor tissues and urine shows that the targeted sequencing of circulating tumor DNA methylation (ctDNAm) can help us
accurately distinguish between HG tumors and LG tumors, MIBC and NMIBC, and is better than genome mutation and fluorescence in situ hybridization (FISH) in terms of recurrence prediction
and prognosis prediction. An article entitled Non invasive diagnosis and survey of
bladdercancer with driver and passenger DNA methodology in a prospective short study published in Clin Transl Med. Now let's go though the detailed research process.

T1DMR and T2DMR DNA methylation of related genes is related to
BLCA occurrence
The differentially methylated region (DMR) of each group can
be obtained by statistically comparing the DNAm from different tissues (see Figure 1 A). The researchers
selected T1DMR (differentially methylated region of normal cells) and T2DMR (differentially methylated region of
cancer cells) for research.
The methylation of cytosine at multiple CpG sites on a single DNA is
called methylation haplotype. In the process of cancer tumorigenesis, the methylation haplotype of T1DMR will not
change, and it will not cause cancer, while the methylation haplotype of T2DMR will change, and it
is closely related to the drive of cancer process and tumor deterioration. Therefore, researchers called T1DMR the
“passenger” and T2DMR the “driver”, and sequenced single-cell RNA and ATAC of 17 T1DMR and T2DMR related to BLCA. The results showed that LG tumor
and NMIBC were only related to the haplotype of T1DMR, while HG tumors and MIBC were related to the haplotype of
T2DMR (Figure 1C). The haplotype statistics of patients with different tumor classifications showed that the
methylation of HOXC9, CCN1, MYOM2, and SOX2 genes in T2DMR group was strongly related to the occurrence
of HG tumors (Figure 1D). The haplotype diversity of HG tumors was higher than that of LG tumors, and the T1DMR
haplotypes in HG and LG tumors were not the same, suggesting that there was obvious epigenetic rearrangement in HG
tumors, and HG/MIBC and LG/NMIB were derived from different cells.
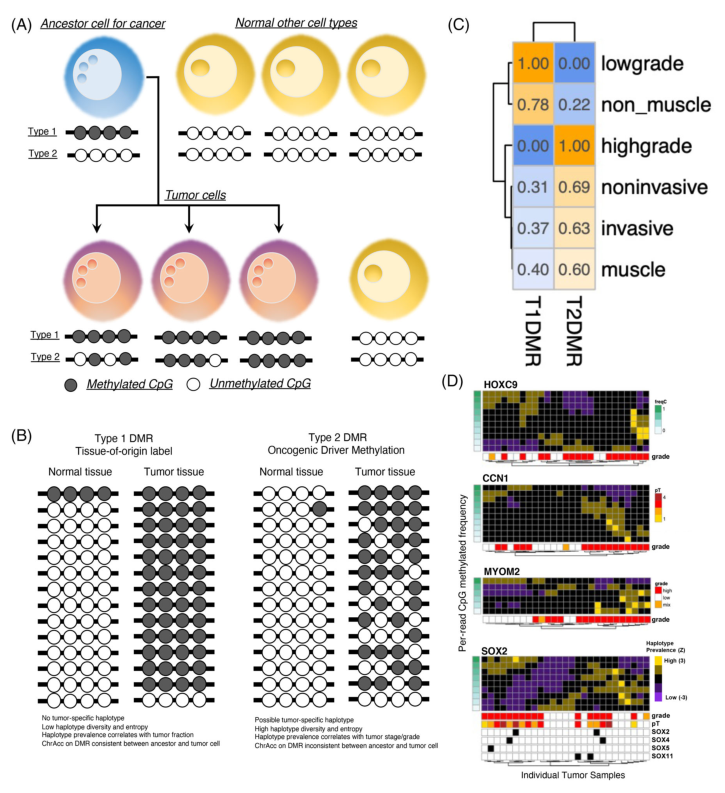
Figure 1. Classification of BLCA differentially
methylated region (DMR)
Construction of SOX2 related T2DMR knockout cell lines to verify
the role of T2DMR in BLCA
The above experiments confirmed that MIBC and NMIBC originated
from the differentiation of different cells. The researchers verified the T2DMR function of SOX2 related driver
genes in MIBC and NMIBC cells (i.e., urothelial basal cells and urothelial mesothelial cells).
The regulatory elements near SOX2 locus and their chromosomal
accessibility were determined through ATAC database. CUT&Tag experiments were performed on BLCA cell lines
T24 (from MIBC donors), 5637 (from MIBC donors) and RT4 (from NMIBC tumors) to study the binding of
transcription factors and chromatin regulatory factors to SOX2 region. The result showed that the combination of
FOXA1 unique to basal cells with the distal regulatory region (L1 or L4) would inhibit the non-enhancer
dependent cis interaction of SOX2 and L4, activate the activity of L1 enhancer, and promote the interaction
between L1 and SOX2 (Figure 2).
In this regard, researchers first used Cas9 lentivirus (provided
by Ubigene) to construct 5637-Cas9, RT4-Cas9 and T24-Cas9 stable cell
lines, and then transduced KO sgRNA lentivirus (provided by Ubigene) of DMR or SOX2 to knockout the SOX2 related T2DMR
region and SOX2 site of the above TM4SF1‐positive cancer subpopulation (TPCS) cell lines to study the regulatory
effect of T2DMR on SOX2 expression. The results showed that after SOX2-T2DMR was knocked out, SOX2 could not
interact with L1 enhancer, resulting in the decrease of SOX2 RNA expression (Figure 2C). The direct knockout of SOX2
will weaken the migration and invasion ability of TPSC cell lines (Figure 3), and has a significant impact on the
invasion of MIBC. These results all indicate that T2DMR is an important regulatory element of SOX2 in BLCA, and its differential methylation will drive the transformation of cancer from NMIBC to MIBC,
suggesting that the differential methylation of T2DMR may be the basis for the classification of MIBC and NMIBC.
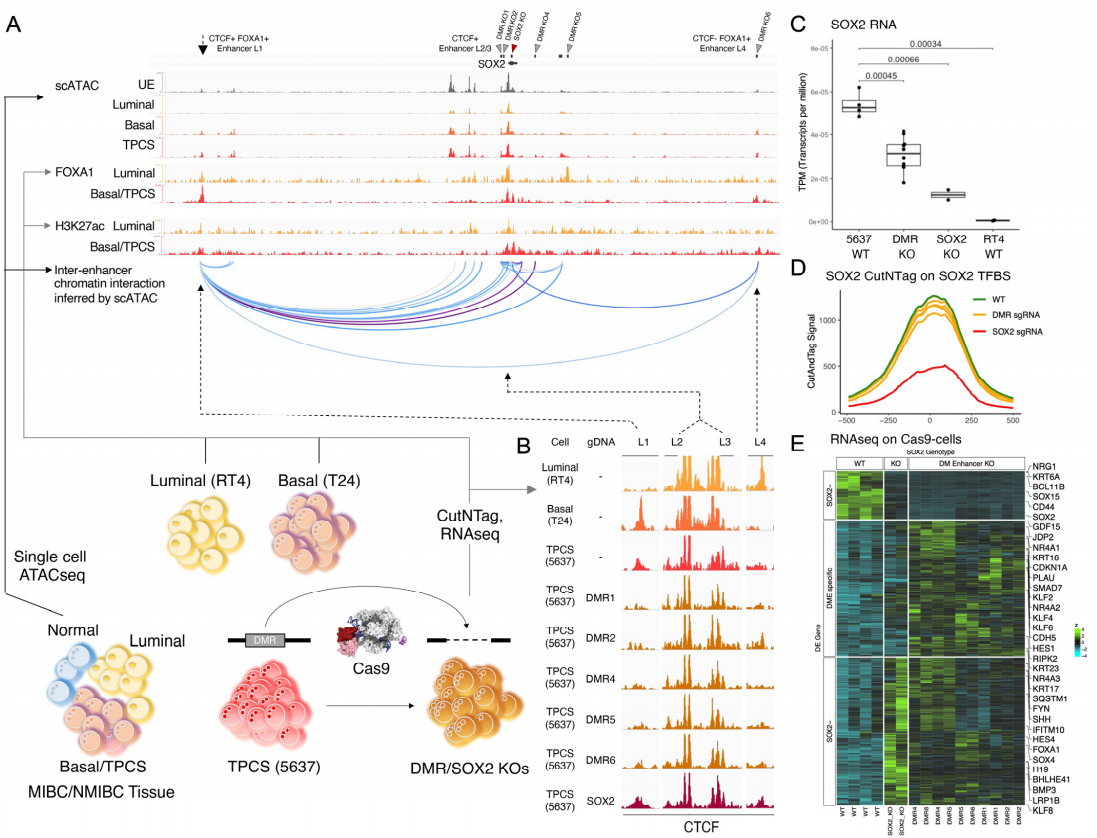
Figure 2. Driver T2DMR interact with oncogene
enhancer to control its function
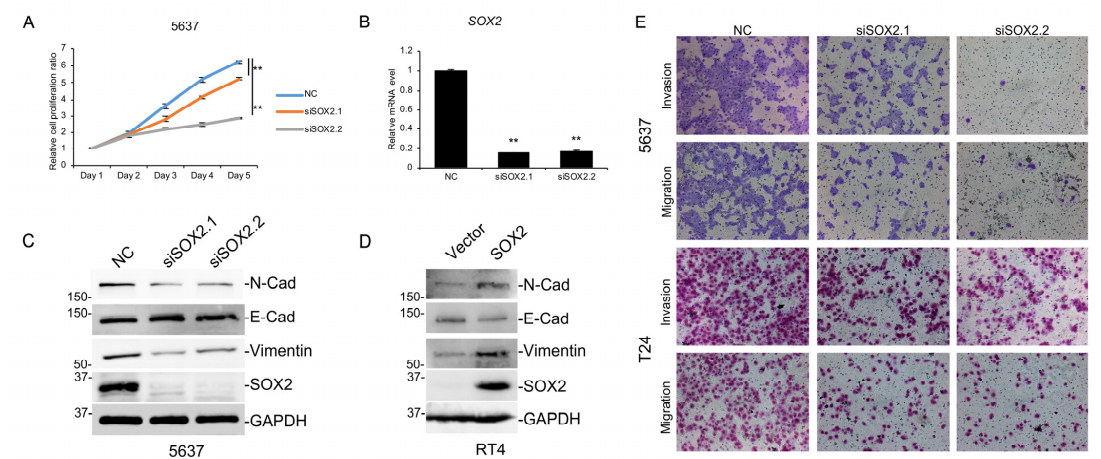
Figure 3 SOX2 function is essential for MIBC
aggressiveness
Ubigene provided in-stock Cas9 lentivirus and the service for
the gRNA lentivirus packaging for this study, assisted in the construction of 5637-Cas9, RT4-Cas9, T24-Cas9
stable cell lines, and accelerated the construction process of SOX2-KO cell line! In addition to in-stock Cas9
lentivirus and lentivirus packaging service, Ubigene also provides adenovirus and AAV packaging services. Click to explore more in-stock lentivirus and custom services>>
10,000+ highly efficient gRNA plasmids are available in stock as low as $20, easily achieve KO and KI. Click to explore our in-stock plasmids>>
Test whether DNA methylation of T1DMR and T2DMR is feasible as the
criteria for accurate classification of BLCA
MIBC and NMIBC tissues were sequenced by EUCAS, and T1DMR and T2DMR DNAm haplotype heat maps were
drawn according to the prevalence (Figure 4A). The heat map shows that the prevalence of DMR haplotype can clearly
classify BLCA samples into HG/LG. In the HG group, there are not only oncogene mutations but also tumor suppressor
gene mutations (TP53, CDKN, SOX), while in the LG group, there are only oncogene mutations (Figure 4A). It was found
that the DNA methylation detection of T1DMR and T2DMR could accurately reflect the potential pathological and
clinical characteristics of tumor biology and be used for accurate classification of MIBC and NMIBC by building
models for prediction (Figure 4B).
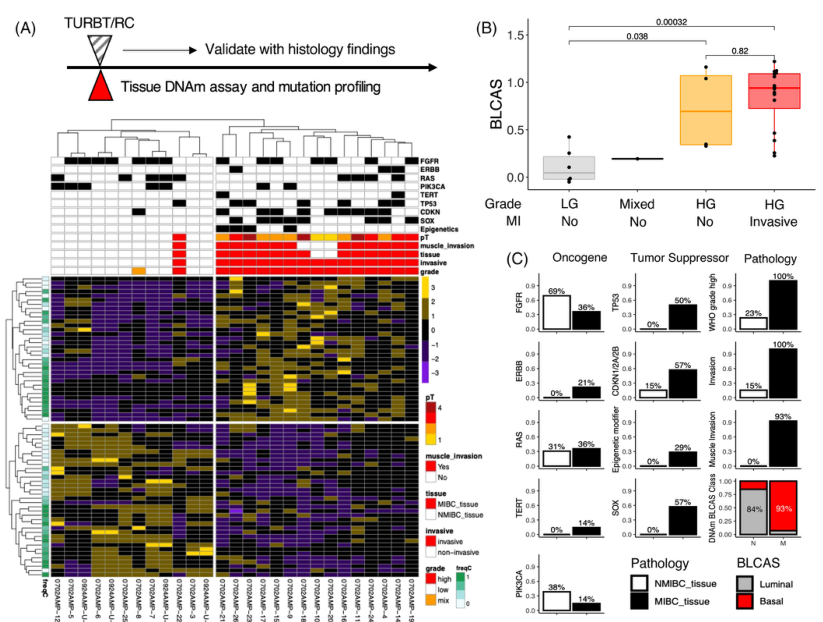
Figure 4. Selective driver and tissue-of-origin DNA
methylation signature outperform tumour genomic mutation in classifying BLCA tumour tissues.
Ubigene provides TP53, CDKN, SOX knockout cell lines to facilitate cancer target validation.
More than 3000 KOs are available in stock, deliver in one week, saving your time in the construction process and
speeding up your research! Only $1780, get high-quality full-allele KO clones. Visit us to
explore your ideal KO>>
Express custom KO cell line service, starting from $2980, full-allele KO clones deliverable. Get a quote>>
Feasibility of
T1DMR and T2DMR related methylation detection in urine for BLCA classification, recurrence and prognosis
prediction
Subsequently, the author tested whether cancer related
methylation characteristics can be recognized in urine, and found that the tumor load was positively correlated
with the DNAm signal in urine. Therefore, the author developed a non-invasive BLCA diagnosis method - to predict
the tumor by multiple PCR NGS sequencing of the DNAm haplotypes in urine samples (UCAS). The authors compared
this method with the traditional BLCA non-invasive detection method, and found that this new method has higher
sensitivity and specificity, and is superior to the existing clinical diagnosis methods.
In addition, the author also confirmed the accuracy of UCAS in
the prediction of HG/LG, MIBC/NMIBC classification by comparing the preoperative and postoperative pathological
results and UCAS detection results. And also confirmed the feasibility and accuracy of UCAS in the detection of
postoperative residual foci and the prediction of recurrence.
This study confirms the utility of DNA methylation on tumor
specific DMR as a biomarker for non-invasive diagnosis of BLCA, and creates a more accurate non-invasive
diagnosis method for BLCA. It is expected to provide more convenient and accurate tumor staging for clinical
treatment of BLCA and ensure the prognosis of BLCA treatment!
Recommended products:
Cas9 stable cell line in stock>> 100+ cell lines, stably express Cas9,
easily achieve gene editing by tranfecting gRNA and donor DNA.
Cas9 lentivirus in stock>>as low as $900, high titer and viability, efficiently transfect
each cell!
Reference:
Xiao, Yu et al. “Non-invasive diagnosis and surveillance of bladder cancer with driver and
passenger DNA methylation in a prospective cohort study.” Clinical and translational medicine vol. 12,8
(2022): e1008. doi:10.1002/ctm2.1008











