IF=17.7|Ubigene's KO cells Inject New Vitality Into Entomological Research

Research Background
It is often said that mothers are great, spending a lot of effort on the growth and
development of
their children. In nature, the vast majority of insects do not exhibit nurturing behaviors. That is, after insect
mothers lay their eggs through oviposition, they quietly pass away. How then do insect mothers meticulously care for
their offspring? Some insects lay their eggs on host plants, providing immediate food upon hatching. Others deposit
eggs in soil or concealed environments to help their offspring survive adverse conditions.
As gregarious insects, locust mothers also prepare their offspring to adapt to high population
densities. When affected by high population densities, locusts alter aspects of their offspring such as egg size,
quantity, cold resistance, and hatching characteristics, etc1-4, creating transgenerational effects induced by
population density. Among these, the synchronous hatching of eggs lays the foundation for the aggregation of
gregarious larvae, adult mating, and large-scale migration. Previous studies have reported that miR-276 in the
ovaries of gregarious locusts induces synchronous hatching of offspring eggs by activating the essential embryonic
development transcription factor Brahma (BRM)3. However, the signal transduction mechanism leading to high
expression of miR-276 in the reproductive system under conditions of high population density remains unknown.
Analyzing the small RNA response mechanism of the reproductive system to external environmental signals will address
key scientific questions that have not yet been answered in transgenerational effect research.
Abstract
Recently, the team led by Dr. Le Kang from the Institute of Zoology, Chinese Academy of Sciences, published a
research paper titled "Parental experiences orchestrate locust egg hatching synchrony by regulating nuclear export
of precursor miRNA" in Nature Communications. In this work, using the population density-dependent
phenotypic plasticity of locusts as a model, they revealed the mechanism by which experiences of high population
density in female locusts regulate synchronous hatching of offspring eggs through the miRNA synthesis pathway in the
reproductive system. Specifically, this study utilized Ubigene -engineered XPO5 knockout 293T cells to assess whether PTBP1 could
function independently of XPO5.
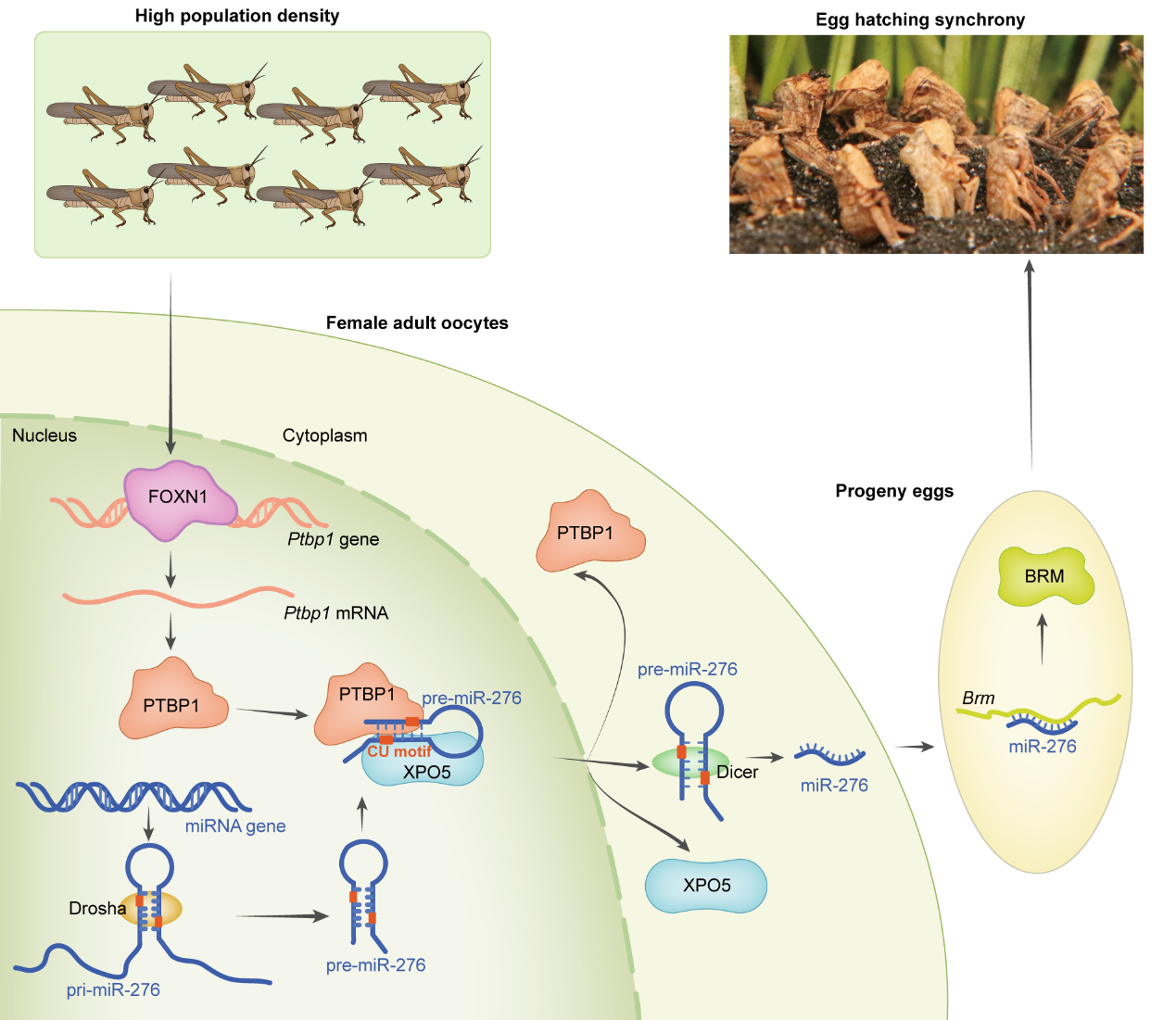
Model of FOXN1-PTBP1–miR-276-regulated egg-hatching synchrony in response to high-density stimuli
Results & Discussion
Firstly, specific primers for intermediate products of miR-276 were
designed as per miRNA biogenesis stages, and the correctness of the sequences of locust miR-276
transcription products (pri-miR-276) and precursors (pre-miR-276) was confirmed in HEK 293T cells lacking endogenous miR-276.
By comparing the expression levels of these intermediates in oocytes of gregarious and solitarious locusts, it was
found that the high expression of miR-276 in the oocytes of gregarious locusts is due to efficient
nuclear export of pre-miR-276 (see Figure 1).
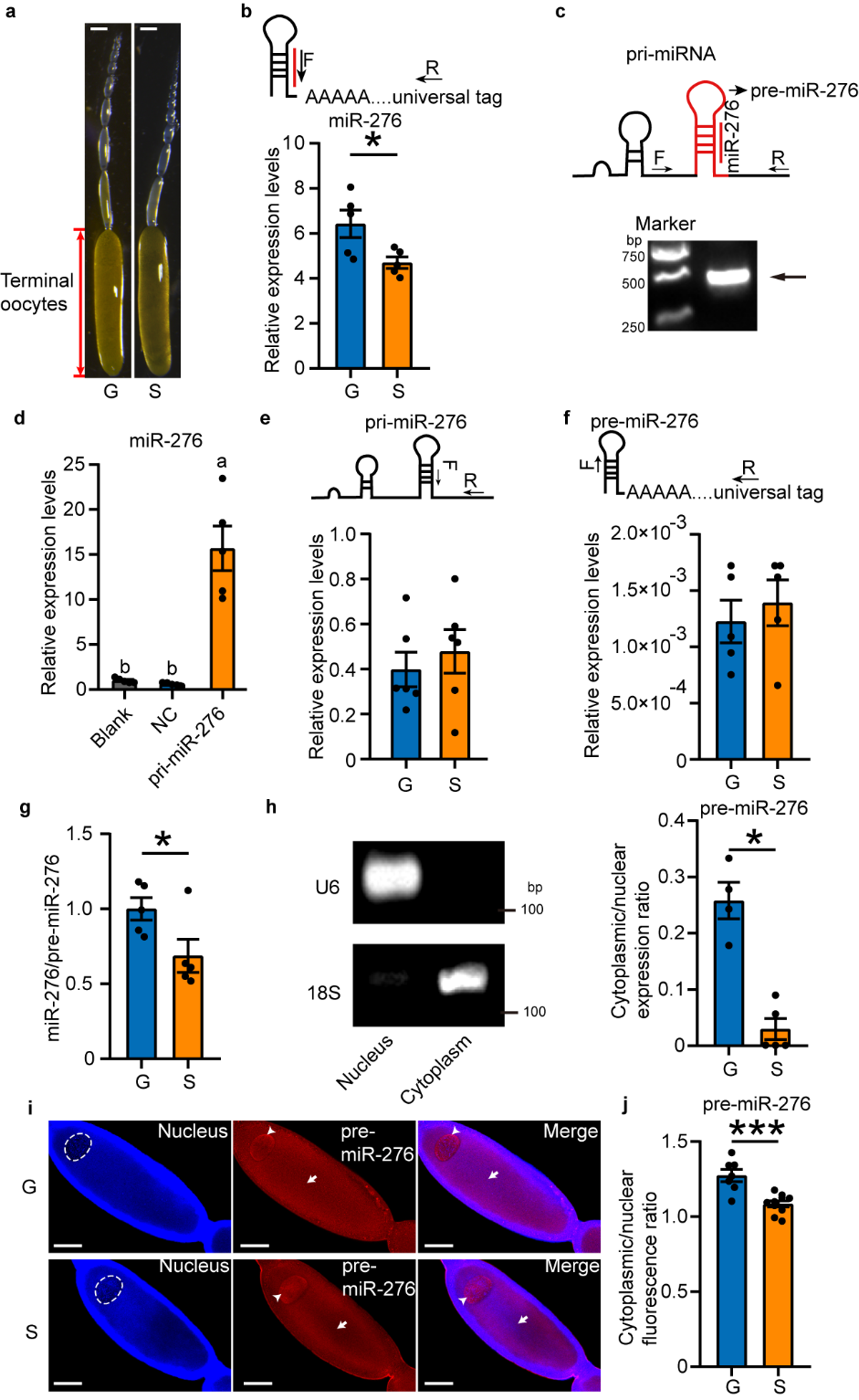
Figure 1 The nuclear export efficiency of pre-miR-276 in terminal oocytes of gregarious locusts is higher than solitarious locusts.
To further explore which protein factors are involved in the
nuclear export of pre-miR-276, researchers conducted RNA pull-down experiments on oocytes using specific probes for
pre-miR-276 to pull out proteins interacting with pre-miR-276, identifying some proteins likely involved in the
nuclear export of pre-miR-276. Combining in vivo interference and in vitro cell overexpression experiments confirmed
that the RNA-binding protein PTBP1 promotes nuclear export of pre-miR-276 (see Figure 2).
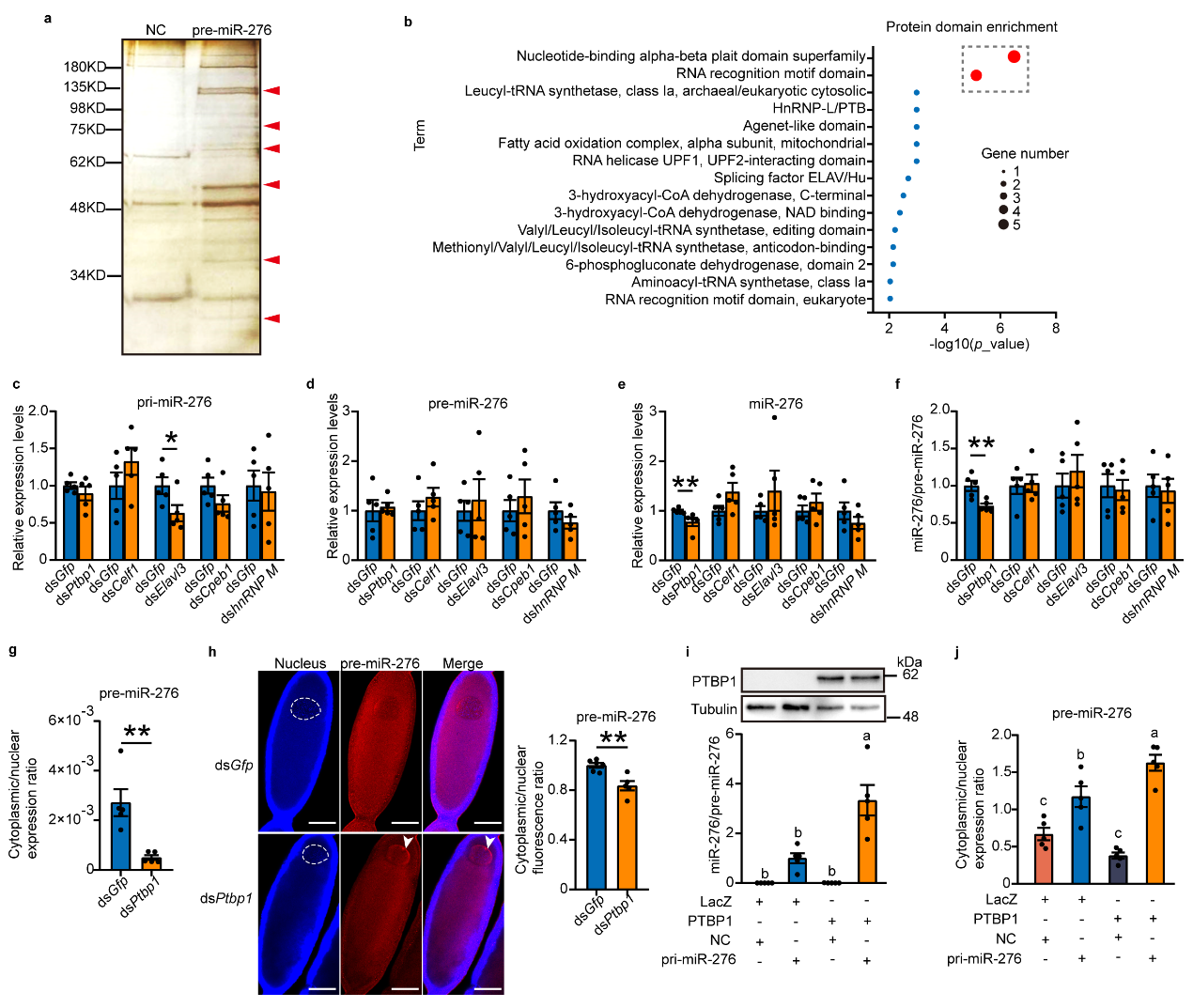
Figure 2 PTBP1 facilitates the nuclear export of pre-miR-276
Since PTBP1 is an RNA-binding protein typically located
downstream of signaling pathways. To investigate how PTBP1 responds to changes in high population density, the study
further searched for upstream factors that respond to population density signals. Through quantitative analysis, it
was found that Ptbp1 in gregarious and solitarious locusts responds transcriptionally to changes in
population density. Through amplification and validation of the Ptbp1 promoter and prediction screening
of 5 potential transcription factors for Ptbp1, followed by in vivo interference, EMSA experiments, and in
vitro cell promoter activity verification, FOXN1 was confirmed as the transcription factor for Ptbp1 (see
Figure 3).
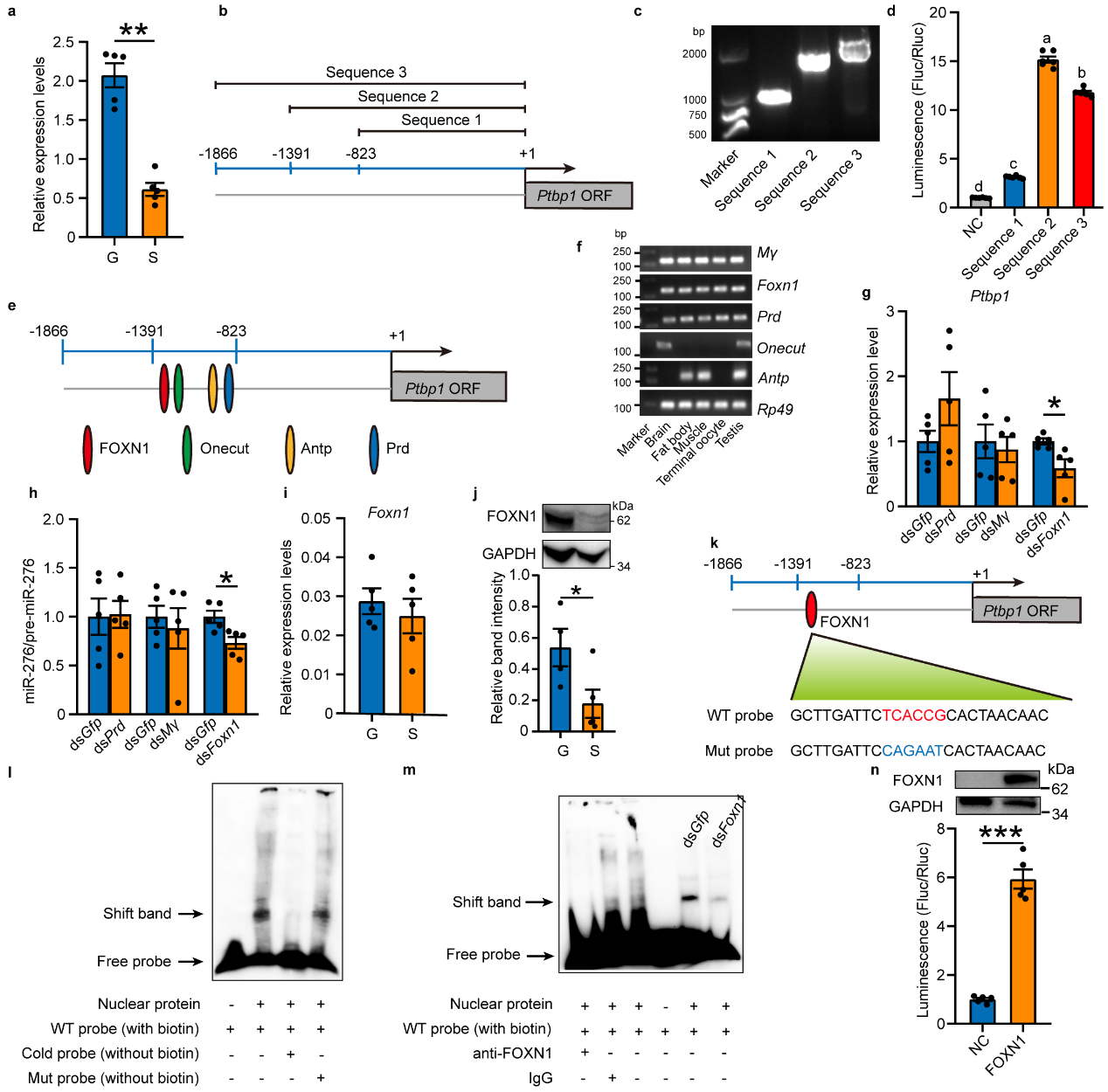
Figure 3 Ptbp1 is transcriptionally activated by FOXN1
To mechanistically study how PTBP1 promotes the nuclear export
of pre-miR-276, this study first confirmed through RNA-protein interaction pull-down experiments that PTBP1 directly
binds to pre-miR-276. Since PTBP1 in locusts is a nucleocytoplasmic shuttle protein, it was hypothesized that PTBP1
might function either independently or in association with the exportin-5 (XPO5) transport protein, which is
necessary for pre-miRNA nuclear export. The study employed Ubigene-engineered XPO5 knockout 293T cells to
evaluate whether PTBP1 could function independently of XPO5. It was found that in the absence of XPO5, PTBP1 cannot
function independently. Through protein interaction modeling predictions and in vivo/in vitro protein interaction
experiments, it was discovered that PTBP1 interacts with XPO5. Importantly, in vivo knockdown and cell
overexpression experiments demonstrated that PTBP1 promotes the nuclear export of pre-miR-276 through its
interaction with XPO5 (see Figure 4).
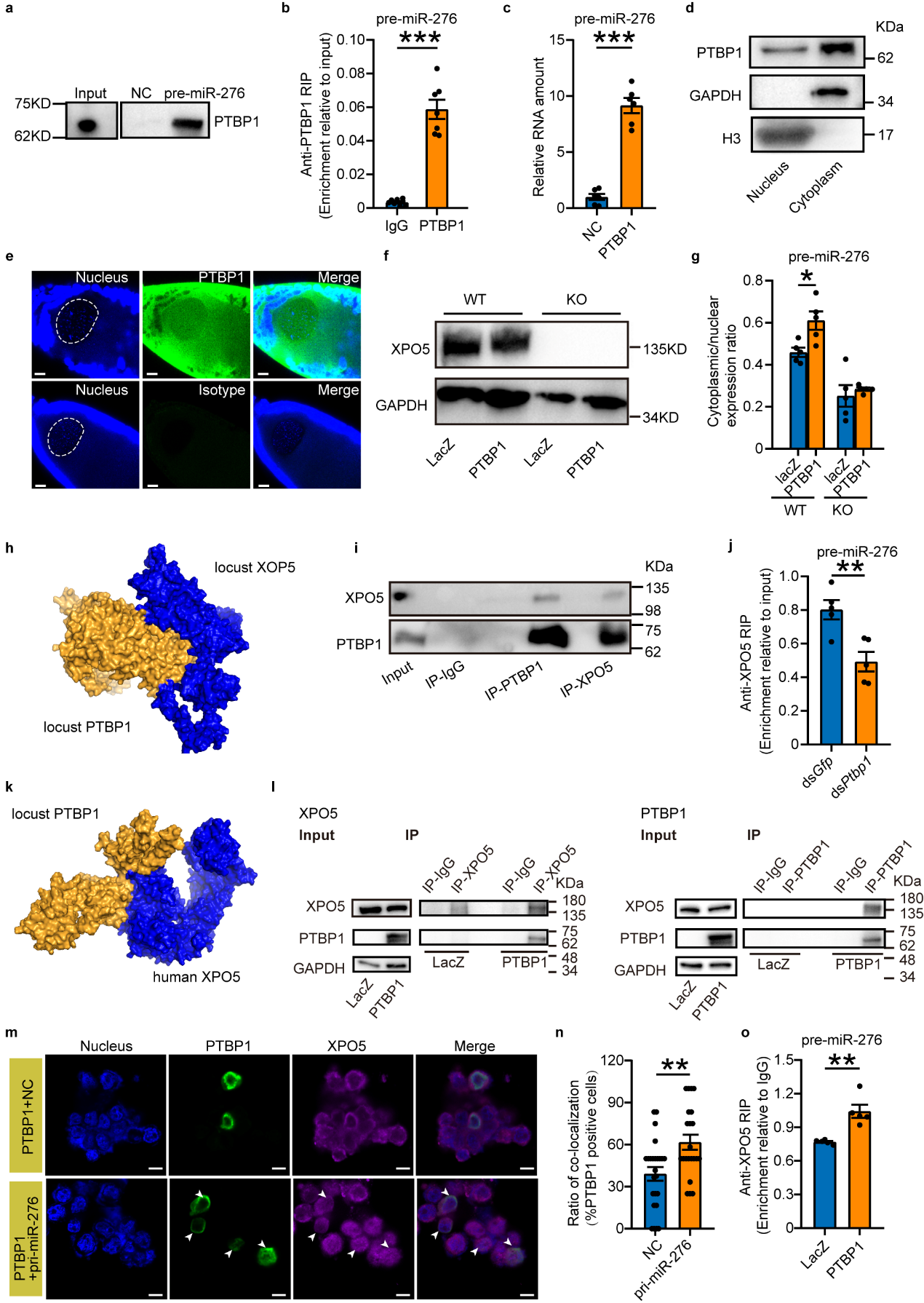
Figure 4 PTBP1 coupled with XPO5 facilitates the nuclear export of pre-miR-276
How can PTBP1 recognize pre-miR-276? This study first speculated
that PTBP1, like XPO5, functions by recognizing the common secondary stem-loop structure present in pre-miR-276.
However, when eight highly expressed miRNAs in the ovaries of gregarious locusts were selected for testing, it
was found that interference with Ptbp1 did not inhibit the nuclear export of these miRNA precursors
(pre-miRNAs). This suggests that PTBP1's role in nuclear export of miRNA precursors is not universal, meaning PTBP1
does not function by recognizing the secondary stem-loop structure of pre-miRNAs. Given that PTBP1 is a
polypyrimidine tract-binding protein6, and the sequence of pre-miR-276 contains a CU motif on both the 5p
and 3p strands, it precisely matches the motif bound by PTBP1. Through designed mutation experiments, it was found
that the promotion of pre-miRNA nuclear export by PTBP1 ceases when either CU motif is mutated. Additionally,
PTBP1's facilitation of pre-miRNA binding with XPO5 is also lost with CU motif mutations, whereas non-CU motif mutations
do not affect PTBP1's promotion of pre-miRNA nuclear export. Therefore, both CU motifs are essential for PTBP1 to
promote the nuclear export of pre-miR-276. Thus, PTBP1 functions to promote the nuclear export of pre-miR-276 by
recognizing these CU motifs (see Figure 5).
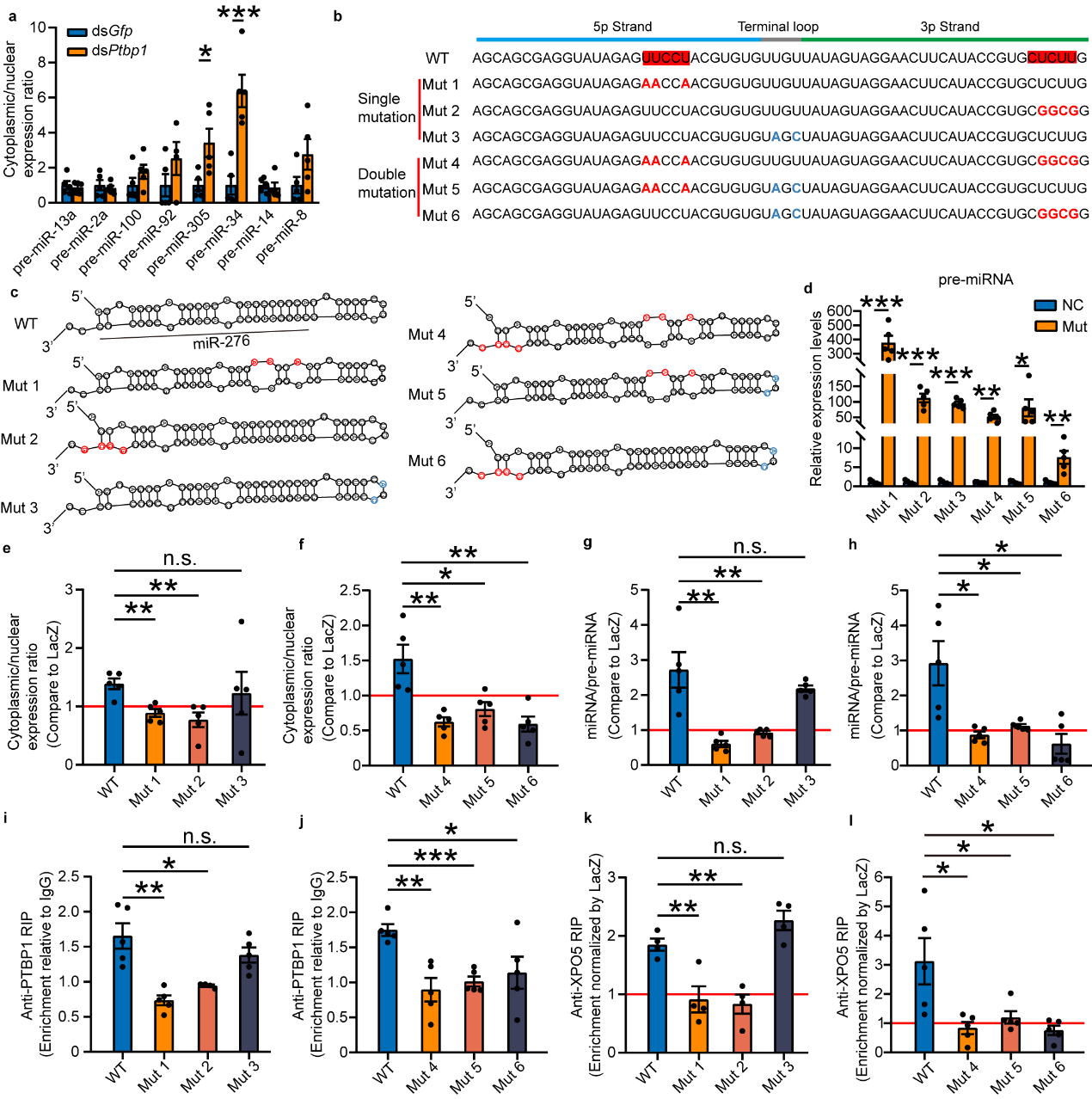
Figure 5 PTBP1-mediated nuclear export is dependent on the "CU motif"
In previous studies, it was found that locusts experiencing high
population density promote synchrony in hatching of offspring eggs by upregulating miR-2763.
Furthermore, the high expression pattern of PTBP1, miR-276, and target BRM in gregarious oocytes
can be transmitted to offspring eggs. To further investigate whether PTBP1 also affects hatching of offspring eggs
by influencing the expression of miR-276, phenotype rescue was carried out by interfering with Ptbp1 in
female insects and overexpressing miR-276 after interfering with Ptbp1. Analysis of hatching time of
offspring eggs showed that PTBP1 in the reproductive system of female insects can promote synchronous hatching of
offspring eggs by activating miR-276 (see Figure 6).
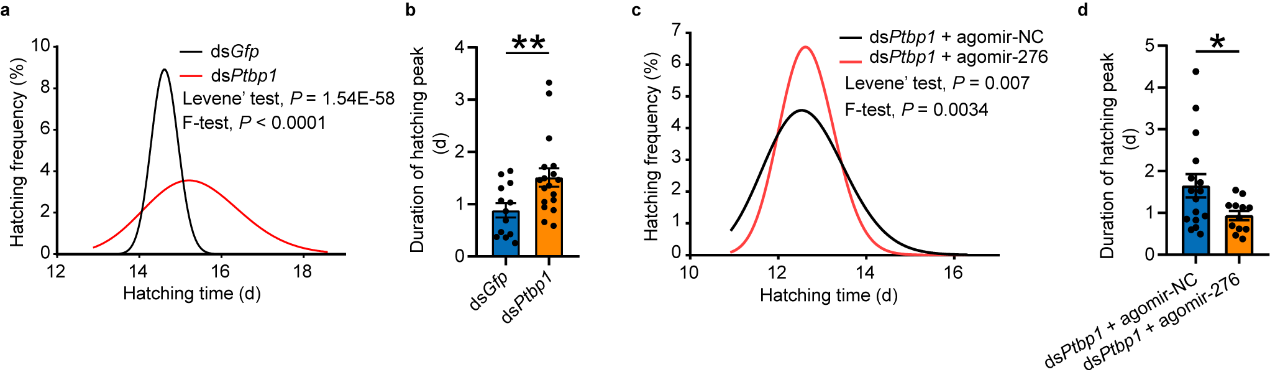
Figure 6 PTBP1–miR-276 regulates egg-hatching synchrony
The study also explored the universality of PTBP1-mediated
nuclear export mechanism of pre-miR-276. Since miR-276 is an insect-specific miRNA, and pre-miR-276, especially the
CU motifs, are highly conserved in insects. Represented by Drosophila, the study verified that PTBP1 can also
promote the nuclear export of pre-miR-276, implying the functional conservation of PTBP1 in insects (see Figure
7).
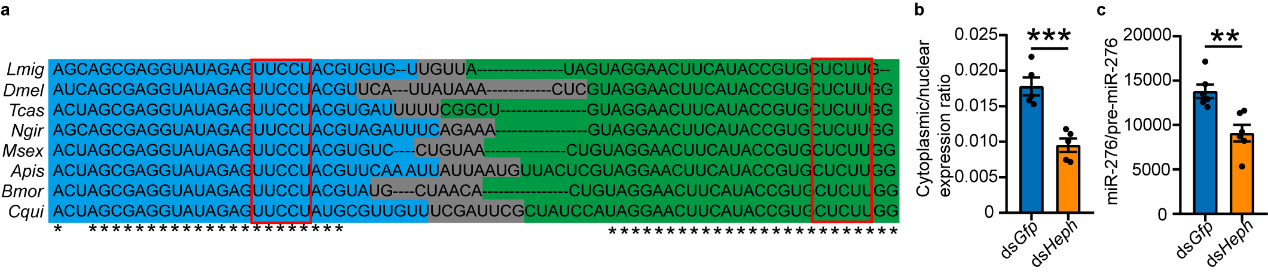
Figure 7 PTBP1 promotes nuclear export of pre-miR-276 in Drosophila
Summary and Prospect
This study reveals that under high population density, the
transcription factor FOXN1 activates PTBP1 in the terminal oocytes of locusts. PTBP1, by recognizing the "tag" on
pre-miR-276, induces its binding to PTBP1 and recruits more XPO5, promoting efficient nuclear export of pre-miR-276
and resulting in high expression of miR-276. miR-276-BRM induction synchronizes offspring egg hatching. Moreover,
PTBP1's mechanism of promoting nuclear export of pre-miR-276 is conserved in insects. This study integrates animal
life experiences with micro-molecular regulatory pathways inside organisms, proposing new insights into miRNA
synthesis processing and responses to the environment. It deepens understanding of the mechanisms of environmentally
mediated transgenerational effects. Specifically, it suggests that locusts regulate synchronized hatching of
offspring eggs based on their life experiences, laying the foundation for future research on larval aggregation and
mass migration of adult locusts. Thus, parental preparation of offspring life history strategies based on their own
life experiences facilitates adaptation of offspring to the environments their mothers once lived in.