[Case study] CRISPR library screen for target candidates -- Essential genes in AGS cell line
Previously we released an article for CRISPR library, which mentioned the application of CRISPR library (Click here to review>>CRISPR library 101 (Part I) what are the popular applications of CRISPR library? >>CRISPR library 101 (Part 2) - how to select and construct a CRISPR library?). And in this article, we will share a typical case of CRISPR library application in drug target screening. Hope this could help you better understand how to utilize CRISPR library in your research!

Gastric cancer is the fifth most common type of cancer in the world, and also the third highest mortality tumor disease. Although great progress has been made in the research of early diagnosis, surgery, chemotherapy and radiotherapy of gastric cancer, the cure survival rate of gastric cancer is still very low, and the survival rate in recent five years is only about 30%. Therefore, it is urgent to find new drug targets and develop better treatment for this cancer.
Zeng et al. used CRISPR knockout library to screen the whole genome of gastric cancer cell (GCC), and found 184 new essential genes, and identified 41 essential genes as potential drug targets. After further research, they confirmed METTL1 as a potential therapeutic target for gastric cancer. The procedures of this research are as follows:
Genome wide screening of AGS cell line using CRISPR library and evaluation of the reliability of the result
By comparing and analyzing the correlation coefficients of GC samples in Achilles and Sanger databases, the researchers obtained seven overlapping GC cell lines (see Figure S1), and found that once the definition of essential genes was changed, the correlation between the seven overlapping GC cell lines' essential gene databases would be reversed: 1969 essential genes defined by Achilles showed strong correlation between the two databases; 525 essential genes defined by Sanger have weak or no correlation between the two databases (see Figure S1C and Figure S2C).
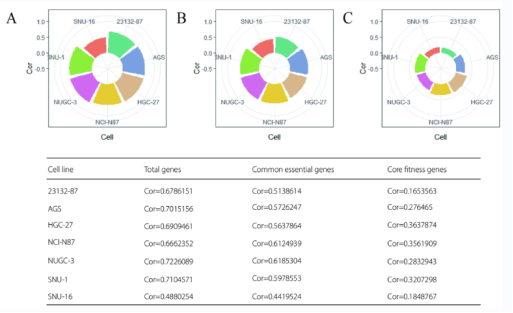
Figure S1
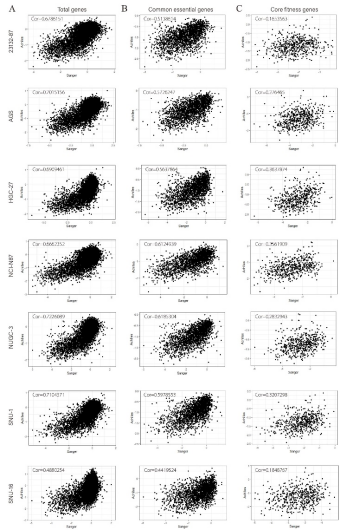
Figure S2
Therefore, the researchers decided to use the whole genome CRISPR library to determining key genes and therapeutic targets of GC cells. TKov3 library transfected into 293T cells by liposome method for virus packaging. Then the AGS cells were infected with the virus, then screened by purinomycin. After extracting the DNA of the control group cells (without any pressure) on day 0, day 10 and day 20 for deep sequencing and comparison, they found that the number of gene deletions increased with the increase of cell passage (see Figure 1C). Combining Wilcoxon rank and the test result, they confirmed that the screening was succeed.
Then, the researchers used MAGeCKFlute algorithm and RRA score to identify 854 essential genes of AGS cell lines, and established a new database for these genes. Through Pearson correlation coefficient analysis, it is found that the genes in this new database are highly correlated with the essential genes in Achilles and Sanger databases, and the area under the curve for evaluating the genes sequenced by RRA score by comparing the essential genes and non essential genes list of Moffat in TKov3 library is 0.993 (Figure 1I). These results indicate that the screening result of CRISPR library is highly reliable.
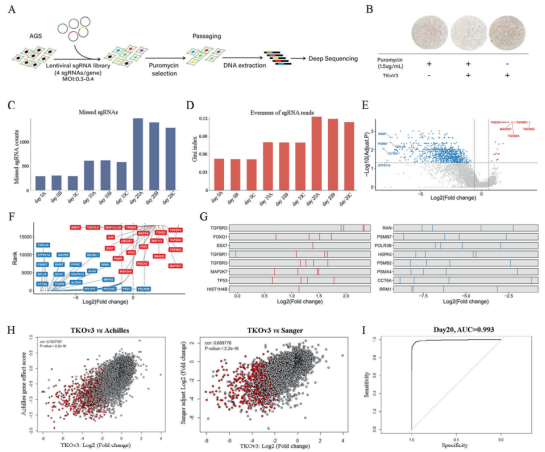
Figure 1. Negative screening of AGS whole genome using CRISPR library
Ubigene focuses on the field of gene editing, has rich experience in cell gene editing, and provide eight in-stock CRISPR libraries and three types of customized libraries (CRISPR-KO, CRISPRa, CRISPRi), providing one-stop solution including high-throughput sgRNA library construction, virus packaging, cell transfection, drug screening, NGS sequencing and data analysis. Library coverage >99%, uniformity< 10. Visit us for more details>>
Explore the function of essential genes
Then, by analyzing the screening results, they found that 689 of the essential genes screened on the 10th and 20th days were the same. Then they used GO biological function enrichment analysis to study the functions of all essential genes at each time point, and found the 10 most closely related functions (Fig. 2B). They found that the analysis result on the 10th day only showed that essential genes were closely related to maintaining normal ribosome and RNA functions, while the analysis results on the 20th day could be specific to heme synthesis process, cellular drug response and other functions (Fig. 2E). This shows that the screening results of CRISPR library are reliable, and the specific functions of essential genes can be found out by prolonging the screening time.
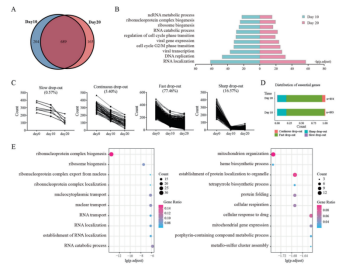
Figure 2. Essential genes screened from TKov3 library at different time points
The researchers analyzed 9 essential genes and 1 nonessential gene in the screening results, and found that only the sgRNA of SNRPB and TOP3A had different effects, and the sgRNA of the other 7 genes had consistent effects on cell activity (Figure 3A). Generate knockdown cell models of these 10 genes in AGS cell line to study the effects of these genes on cell activity (Fig. 3B). ALamar Blue test results show that knockdown of non essential gene RNASET2 will not affect cell activity and colony formation, while knockdown of essential genes will reduce cell activity and number of cell colonies (Figure 3D, E), which also verifies that the screened genes are indeed necessary for the survival of AGS cells.
Among the 9 essential genes, CIT and TACC3 were not previously included in Achilles and Sanger databases as essential genes, but they are indeed closely related to maintaining cell activity. There are other 182 such genes from TKov3 library screening results. Therefore, these 184 newly discovered essential genes fill the gaps in Achilles and Sanger databases. Moreover, the researchers also found that compared with normal gastric mucosa tissues in TCGA database, the expression of 9 genes in gastric adenocarcinoma (GAC) tissues was up-regulated (Figure 3G), suggesting that the essential genes screened from the library may be the potential drug targets for cancer treatment.
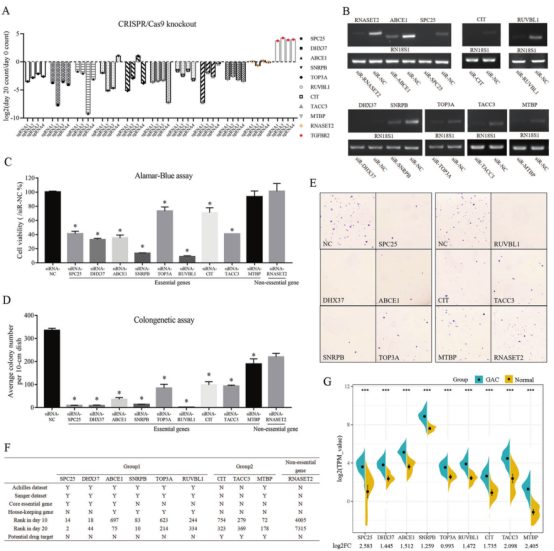
Figure 3. Evaluation of 9 essential genes
Ubigene has 3000+ KO cell lines in stock, covering 27 signal pathways, 26 diseases, and 5 categories of drug targets, Starting from $1980. 1 week's express delivery. Homo knockout clones. To explore our KO cell bank>>
We also offer custom KO cell line service, starting from $2980. Moreover, shRNA mediated knockdown stable cell line only $1780. Get a quote>>
Screening potential drug targets from essential genesIn order to screen potential drug targets with high specificity, researchers first removed essential genes and only retained the essential genes specific for AGS cells. Then, PMA MAS 5.0 was used to remove the genes with low TCGA-STAD expression in AGS cells, and the remaining TCGA-STAD differentially expressed genes were compared. It was confirmed that 41 essential genes up-regulated in GAC tissues were potential drug targets (Figure 4A), including CIT, TACC3, and MTBP mentioned above.
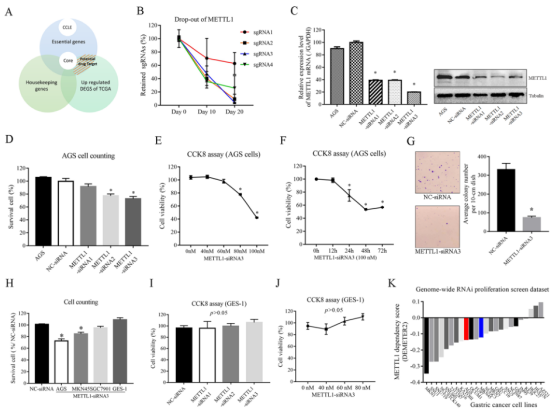
Figure 4. Screening of potential drug targets
Knockout METTL1 in AGS cells to verify the effectiveness of screening strategy
Like CIT and TACC3, tRNA N7-m7G methyltransferase (METTL1) gene was not included as an essential gene by Achilles and Sanger, but was identified as an essential gene in CRISPR library screening. To further verify the effect of METTL1 on the biological function of GC cell lines, researchers designed three siRNAs targeting METTL1 and established METTL1 AGS knockdown cell line. Knockdown of METTL1-siRNA2 and METTL1-siRNA3 led to a 22.46% decrease in cell survival rate and a 27.09% decrease in cell number (Fig. 4D). CCK8 experiment results showed that cell viability decreased in a dose and time-dependent manner (Fig. 4E, F). Knockdown of METTL1 also led to a 77.82% decrease in cell colony number (Fig. 4G) in colony formation assay. These all indicate that METTL1 gene is crucial to the survival of AGS cells and is an essential gene for AGS cells. This conclusion is consistent with the previous experimental results, which positively demonstrates the effectiveness of CRISPR library screening strategy.
Then, the researchers used the same method to study the role of METTL1 in other GAC cell lines, and found that METTL1 may affect the proliferation of multiple GAC cell lines (Fig. 4K).
Conduct METTL1 knockdown test in vivo to verify whether METTL1 is a potential drug target
MKN45 cells transfected with METTL1-siRNA or NC-siRNA were subcutaneously injected into BALB/c nude mice to form subcutaneous tumors. The volume and weight of tumors were measured on the 20th day. It was found that the average volume and weight of tumors in METTL1 knockdown group were 55.19% and 55.97% lower than those in the control group. It shows that knockdown METTL1 can effectively inhibit tumor growth, and METTL1 can be a drug target for gastric cancer treatment.
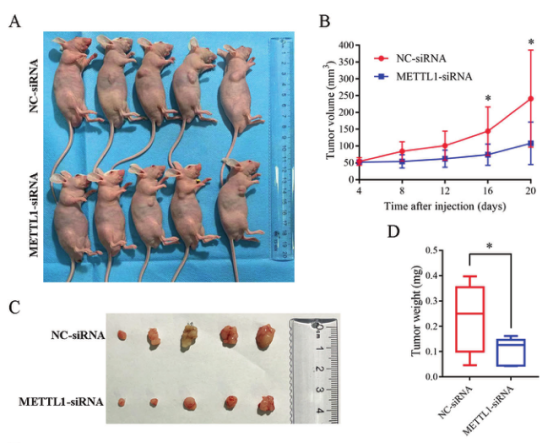
Figure 5. METTL1 knockdown can inhibit tumor growth in vivo
Explore the role and mechanism of METTL1 in GC cellsThe cell cycle of GC cells with METTL1 knockdown was studied. It was found that the level of cell cycle marker CDK1 decreased significantly after METTL1 knockdown. Further studies found that knockdown of METTL1 significantly decreased AKT and STAT3 levels (Figure 6), suggesting that METTL1 may play a role in cell cycle by activating AKT/STAT3/CDK1 signaling pathway.
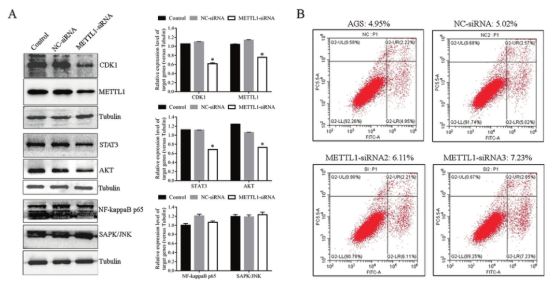
Figure 6. Analysis of the role of METTL1 in AGS cell line
Finally, the researchers also analyzed the clinical sample data in GEO and TCGA databases, and found that METTL1 mRNA in GAC tissues was indeed overexpressed compared with normal gastric mucosa tissues, and the expression of METTL1 mRNA gradually increased with the increase of tumor stage, which indicates that METTL1 may be an effective anti-tumor drug target.
This study found 184 newly established essential genes of AGS cell line, identified 41 potential drug targets, verified the role of METTL1 in the development of tumor cells, and presented a complete screening procedure of CRISPR library: from infecting cells with CRISPR gRNA library, cell screening, DNA sequencing to gene function validation.
Ubigene provides eight in-stock CRISPR libraries, including the whole genome (human/mouse), kinase, nuclear protein, metabolic genes, etc. It also provides on-stop solution from plasmid library construction, virus packaging, to cell screening and NGS sequencing analysis. We also offer CRISPR-KO, CRISPRi, and CRISPRa library construction services to meet different needs. Talk to our technical team for more details>>















