New Approach to Screening Viral Restriction Factors Using CRISPR Libraries!

In Vol.1, we introduced an approach to screen for viral restriction factors using CRISPR library technology, but this screening approach had certain limitations. Today, Ubigene will introduce another approach for screening viral restriction factors using CRISPR libraries. Let's take a look together.
Background
HIV-1 is the pathogen responsible for the AIDS pandemic, posing a severe challenge to global public health for over 40 years. Viral restriction factors not only inhibit HIV-1 infection but also have broad effects on other viral pathogens, and often play roles in inflammation and cancer. Therefore, the search for new viral restriction factors is highly attractive to researchers. Based on CRISPR library screening technology, researchers have discovered numerous genes related to viral infection and replication, providing insights into virus-host interactions. However, these screenings mainly identify differential genes by enriching sgRNAs in cells that become more sensitive or resistant to the virus after infection; the screened genes tend to be more related to cell survival and proliferation rather than viral infection.
To address this issue, researchers combined CRISPR/Cas9 library technology with selectable and replicable HIV-1 viruses. By detecting the abundance of sgRNAs carried in viruses released into the culture supernatant after multiple rounds of continuous viral infection of cells, they could more directly screen for genes related to viral infection and replication.
Abstract
Innate antiviral factors are key to effectively defending against viral pathogens. Current methods for discovering antiviral factors typically focus on the initial stages of viral replication and are mostly limited to single-round infections. Prelli Bozzo et al. designed a CRISPR knockout library targeting 511 genes potentially related to HIV virus infection, reverse transcription, replication, and other biological processes, and integrated these sgRNAs into replication-competent HIV viruses, termed the "traitor virus" library. Using HIV virus to infect CEM-M7 cells and collecting cell supernatants at different time points, they detected the abundance of sgRNAs in the supernatant viruses through NGS sequencing. The study found that as the infection time increased, sgRNAs targeting GRN, CIITA, EHMT2, CEACAM3, CC2D1B, and RHOA were enriched over 50-fold in the viruses from the cell culture supernatant.
Discovering Viral Restriction Factors with CRISPR Libraries
Library Type: Custom CRISPR knockout library
Transduced Cells: CEM-M7 cells
Virus: HIV-1 (NL4-3 and CH077)
Screening Strategy: Collect virus supernatant at different time points, positive selection
Screening Method: Integrate sgRNA sequences targeting different genes into the HIV-1 viral genome. After infecting Cas9 stable expression CEM-M7 cell lines with the virus, the sgRNAs carried in the virus edit the genes in the target cells, thereby affecting the efficiency of viral infection, proliferation, or release. Continuously culture the virus-transfected cells and collect virus supernatant every five days, for a total of 4 collections; during this period, viruses with proliferation advantages will be enriched in the supernatant. After amplifying the sgRNA information carried in the supernatant viruses, use high-throughput sequencing technology to obtain read information of different sgRNAs carried in the viruses. Utilize bioinformatics analysis tools to identify enriched sgRNA sequences, thereby determining candidate antiviral factors/genes.
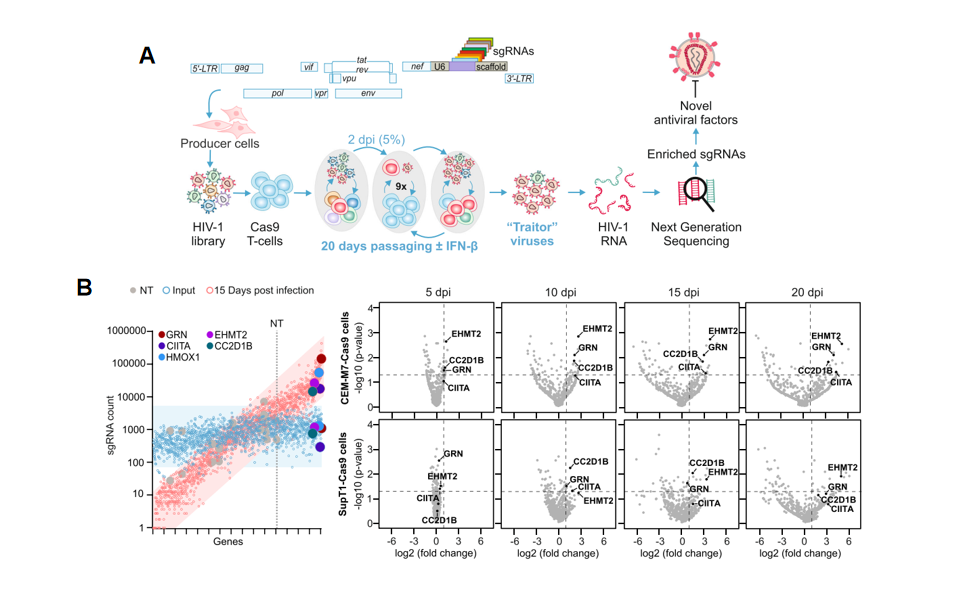
Figure 1 "Traitor virus" library screening process and results
Based on the above experiments, researchers found that in the "traitor virus" library constructed using the NL4-3 virus, the proportion of GRN, CIITA, EHMT2, CEACAM3, CC2D1B, and HMOX1 genes in the virus supernatant increased over time. This indicates that these genes are potential antiviral factors, and when these genes are knocked out, the proliferation ability of HIV-1 virus increases. Subsequently, the researchers conducted experiments on these genes separately to elucidate which stages of virus inhibition these antiviral factors act on.
GRN, CIITA, and CEACAM3 Genes Inhibit Viral Replication in CEM-M7 Cells
After knocking out GRN, CIITA, and CEACAM3 genes in CEM-M7 cells using the CRISPR/Cas9 system, they studied which aspects of viral replication each gene affects. They found that the GRN gene directly inhibits viral replication, while CIITA and CC2D1B genes inhibit viral replication by suppressing viral transcription and release.
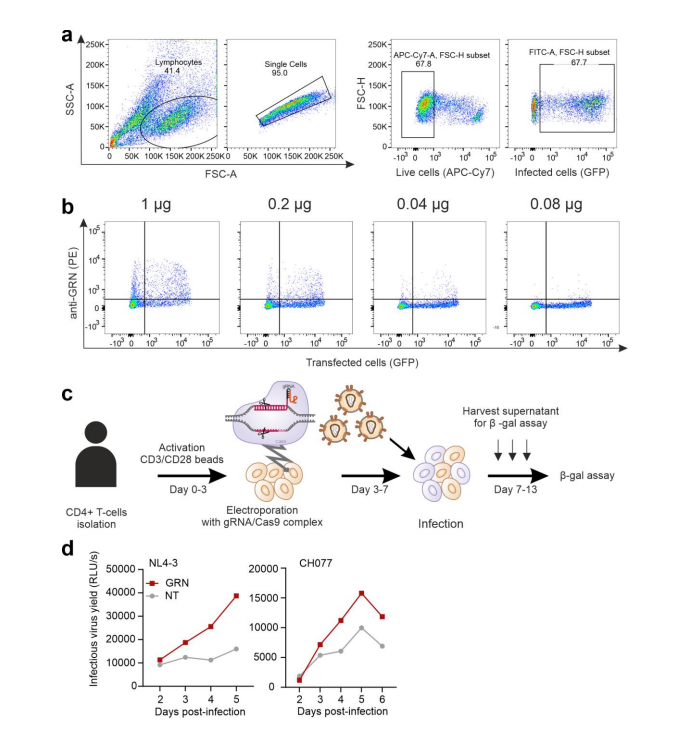
Figure 2 GRN gene knockout increases viral replication
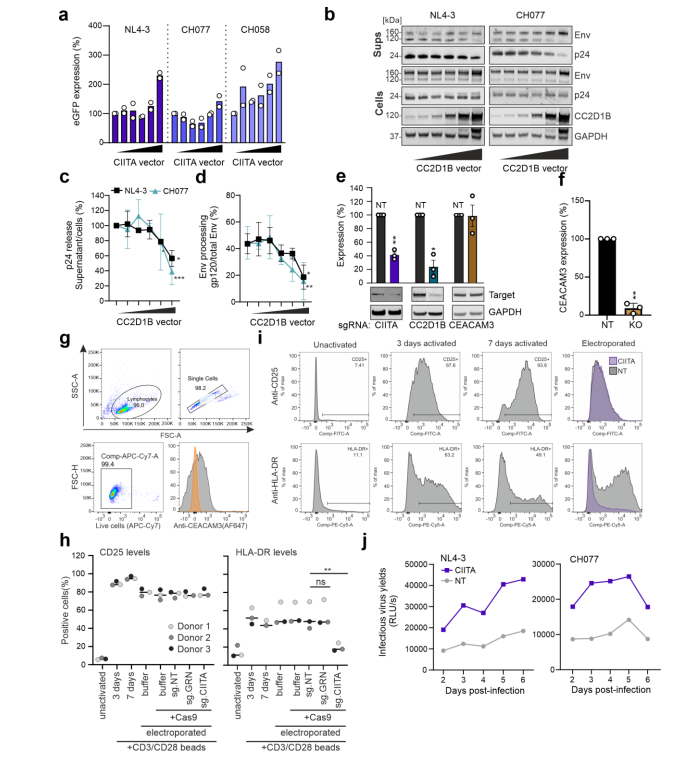
Figure 3 CIITA and CC2D1B genes inhibit viral transcription and release
Screening for Viral Restriction Factors Using the CH077 "Traitor Virus" Library
The study initially chose the NL4-3 virus strain because it is widely used in most research. However, this strain can efficiently replicate in T cells, which might prevent the identification of viral restriction factors that inhibit viral replication through the NL4-3 constructed "traitor virus" library. Therefore, the researchers switched to using the CH077 virus strain to construct the "traitor virus" library for screening. The results showed that among the genes screened by the CH077 "traitor virus" library, CEACAM3, CC2D1B, EHMT2, and HMOX1 genes overlapped with the NL4-3 "traitor virus" library screening results, while the RHOA gene was only enriched in the CH077 "traitor virus" library screening. The researchers further validated the RHOA gene separately through gene knockout.
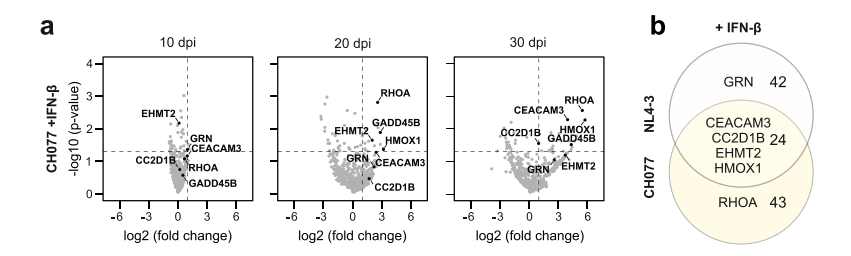
Figure 4 "Traitor virus" CH077 library screening results
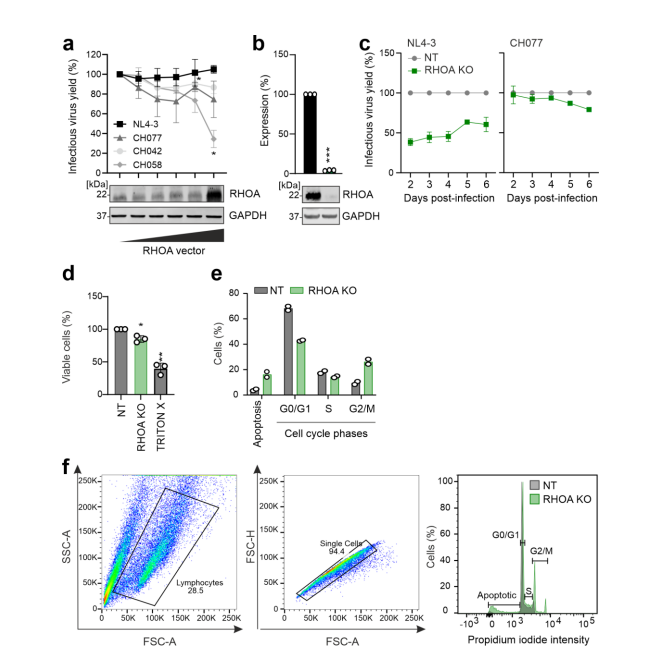
Figure 5 RHOA gene knockout affects viral replication and inhibits cell cycle
Conclusion
Unlike conventional CRISPR library screening—which obtains target genes by enriching sgRNA information in the remaining cells at the end of screening—this study constructed a "traitor virus" library carrying sgRNA information, then identified target genes by collecting virus particles released in cell supernatants, combined with NGS sequencing methods. Through this method, researchers discovered multiple viral restriction factors that not only act on viral replication but also on transcription, infection, release, and other stages, demonstrating the effectiveness of this technology in screening for viral restriction factors.
Ubigene offers 35 types of library with corresponding plasmids and virus particles available in stock and also offers library-transferred cell pools. Now Library Cell Pool starting from US$2290 | Virus Particles starting from US$1845. Screen as you wish! Ubigene can also provide one-stop screening services for custom CRISPR libraries, with multiple deliverables to meet different scientific research needs.
References
Prelli Bozzo, C., Laliberté, A., De Luna, A. et al. Replication competent HIV-guided CRISPR screen identifies antiviral factors including targets of the accessory protein Nef. Nat Commun 15, 3813 (2024).












