Published on: June 18, 2024
Views:
--
CRISPR Library | Unlocking New Insights into Breast Cancer Targets

Breast cancer is the most commonly diagnosed malignancy in women, and it is the second leading cause of cancer-related death in women worldwide[1]. Research and advancements in its treatment have always been a focus in the medical field. According to the expression status of receptor molecules in tumor tissue, breast cancer can be classified into: Luminal A, Luminal B, HER-2 overexpressing, and Triple-Negative Breast Cancer (TNBC)[2]. Notably, triple-negative breast cancer does not express the typical hormone receptors or HER-2, making traditional endocrine therapy and HER-2 targeted therapy less effective in these patients[3]. Therefore, a deeper understanding of breast cancer is needed to develop new treatment methods, providing breast cancer patients with new treatment options and hope.
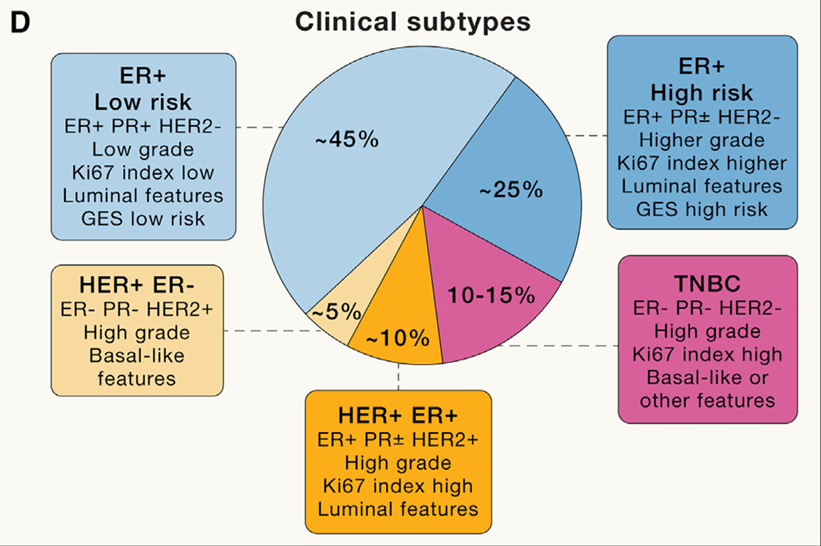
Figure 1: Subtypes of Breast Cancer and Their Distribution[2]
With the rapid development of gene-editing technology, the CRISPR-Cas9 system has brought revolutionary changes to cancer research due to its remarkable precision and flexibility. The CRISPR library, as an application of this technology, enables large-scale screening of gene functions and provides a powerful tool for breast cancer research. Join us in exploring how CRISPR library screening can aid in breast cancer research. We may gain new insights and perspectives and therefore effectively apply this powerful tool!
Discovery of New Targets for Breast Cancer Drug Resistance - NUMEN
The BRCA1 gene is highly associated with hereditary breast cancer, and its loss of function leads to DNA double-strand break repair defects, making cells particularly sensitive to PARP inhibitors (PARPi). However, it has been observed that tumor cells may develop resistance to PARPi[4]. Chen Bohong et al.[5]discovered the gene NUMEN through a whole-genome CRISPR library screen, which is crucial for PARPi resistance in BRCA1-deficient breast cancer cells. NUMEN, through its structure-specific endonuclease activity, promotes non-homologous end joining (NHEJ)-dependent DNA double-strand break repair. This study not only revealed the role of NUMEN in DNA repair and maintenance of genomic stability but also provided potential targets for developing new therapeutic strategies for PARPi-resistant breast cancer.
Library type: Whole-genome CRISPR knockout library (GeCKOv2 library)
Tranduced cell: BRCA1−/− gene knockout MDA-MB-231 cell line with inducible expression of Cas9
Screening type: In vitro drug screening
Screening method: Culture cells under doxycycline condition to induce Cas9 expression, and then add the PARP inhibitor (olaparib) for screening
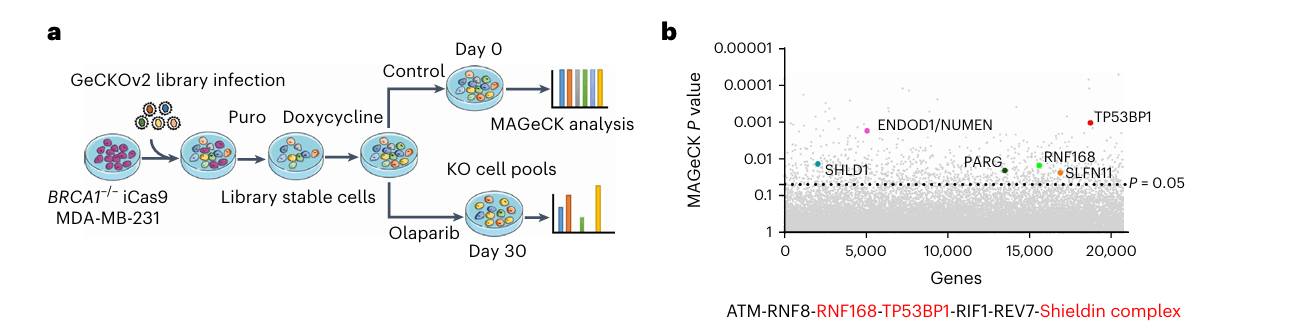
Figure 2: Workflow of Whole-Genome CRISPR Screen[5]
Discovery of Triple-Negative Breast Cancer Targets - Cop1
Triple-negative breast cancer (TNBC) is a cancer type with a poor prognosis and limited response to immune checkpoint blockade (ICB) therapy. The immune cells in the tumor microenvironment (TME), especially the Tumor-associated macrophages (TAMs), play a vital role in promoting tumor growth and metastasis. Wang Xiaoqing et al.[6] identified E3 ubiquitin ligase Cop1 as an important gene that regulates macrophage infiltration and the response to anti-PD-1 therapy through two rounds of CRISPR library screening. Deletion of Cop1 can reduce the infiltration of tumor-associated macrophages, enhance anti-tumor immune responses, and improve the response to ICB therapy for TNBC. Inhibiting Cop1 can improve the effectiveness of cancer immunotherapy for TNBC, providing a potential target for developing new cancer treatment strategies.
Library type: Gene knockout sub-library MusCK 1.0, Gene knockout sub-library MusCK 2.0
Targeted genes: MusCK 1.0: 4500+ genes related to tumor initiation, progression, and immune regulation; MusCK 2.0: 79 candidate genes identified in the first round of screening
Tranduced cell: 4T1 cell line expressing ovalbumin (mOva)
Screening type: In vivo screening of mice, secondary screening
Screening method: To simulate different immune microenvironments, researchers used mice with different immune states, including Foxn1nu/nu mice (immunodeficient), wild-type mice (with normal immune function), and mice vaccinated with ovalbumin (OVA) vaccine. Inject the transduced 4T1 cells into the breast fat pad of mice, and collect tumor tissue at a certain time point after tumor implantation. Based on the 79 candidate genes obtained from the first screening sequencing analysis, the sgRNA library was reconstructed and screening was performed similar to the first screening, and a mouse group vaccinated with OVA vaccine and treated with PD-1 blockade was added.
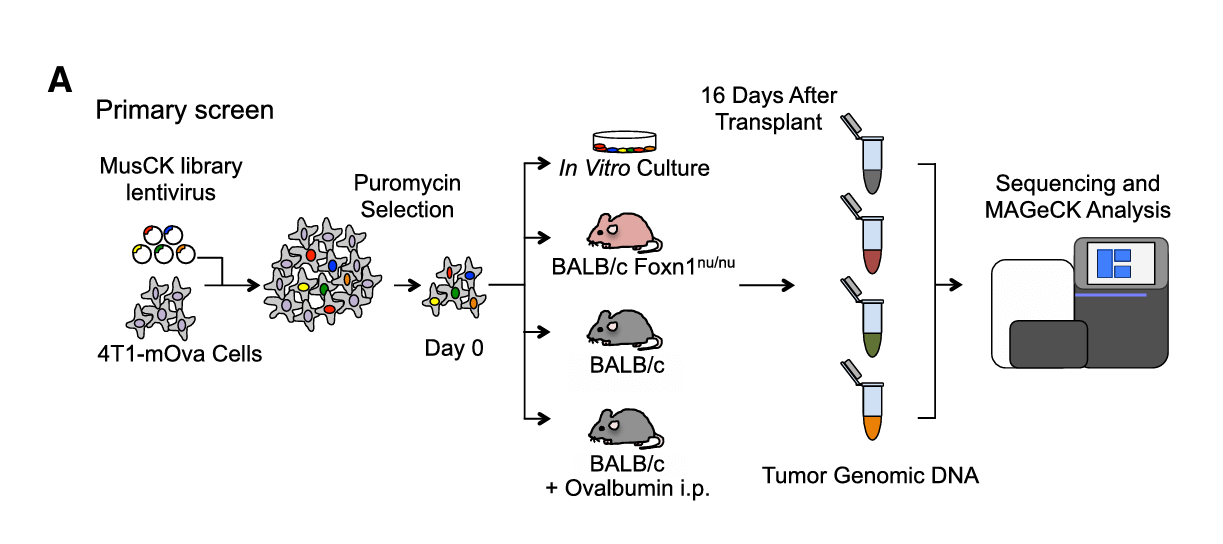
Figure 3: Workflow of the First In Vivo Screening[6]
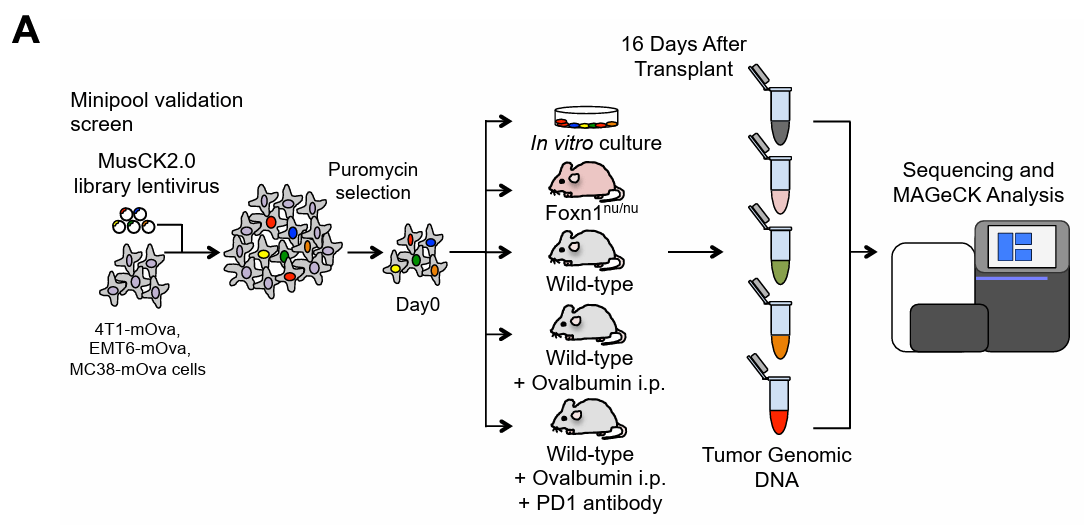
Figure 4: Workflow of the Second In Vivo Screening[6]
New Platform for Identification of Breast Cancer Risk Genes - CRISPR Library
Natasha K. Tuano et al.[7] identified over 200 genes associated with breast cancer risk through a genome-wide association analysis (GWAS). Most candidate pathogenic mutations are located in the non-coding region, affecting cancer phenotypes by regulating gene expression. However, accurately pinpointing target genes and identifying their phenotypes is a challenging task. To overcome this challenge, they conducted screening using 60 CRISPR libraries, ultimately identifying 20 high-confidence genes. These genes promote cancer initiation and progression by enhancing breast cell proliferation or regulating DNA damage response. This study not only demonstrated the capability of CRISPR library screening in accurately identifying risk loci genes but also proved its effectiveness, providing an essential platform for identifying and increasing breast cancer risk-related gene targets.
Library type: Knockout library, inihibition library, and activation library
Targeted genes: 200+ genes associated with breast cancer risk through a genome-wide association analysis (GWAS)
Tranduced cell: 6 different breast cancer cell lines, including K5+/K19+, K5+/K19-, B80-T17, mesHMLE, B80-T5, HMLE
Screening type: In vitro phenotype screening, in vivo mouse screening, in vitro drug synthetic lethal screening
Screening method: 2D and 3D cell proliferation screening: Evaluate the proliferative capacity of cells under two-dimensional (2D) and three-dimensional (3D) culture conditions to simulate cell growth in vivo.
In vivo screening: Inject cells into immunodeficient mice and observe tumor formation.
DNA damage response screening: Treat cells with the PARP inhibitor olaparib to screen for genes that impact the DNA damage response.
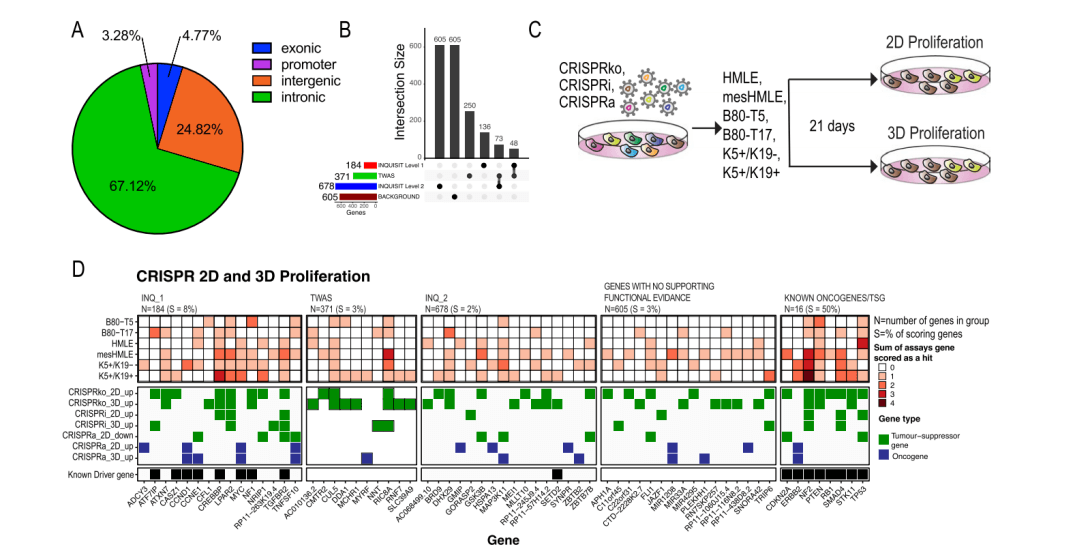
Figure 5: CRISPR Library Screening Identified Breast Cancer Risk Genes that Regulate Proliferation in 2D and 3D Cell Culture Models [7]
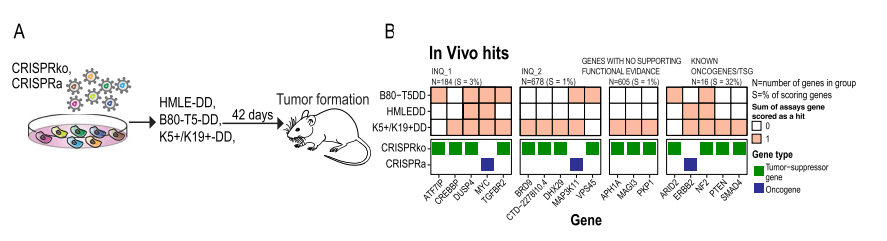
Figure 6: CRISPR Activation and Inhibition Screening Identified Breast Cancer Risk Genes that Regulate Tumor Growth in Mice [7]
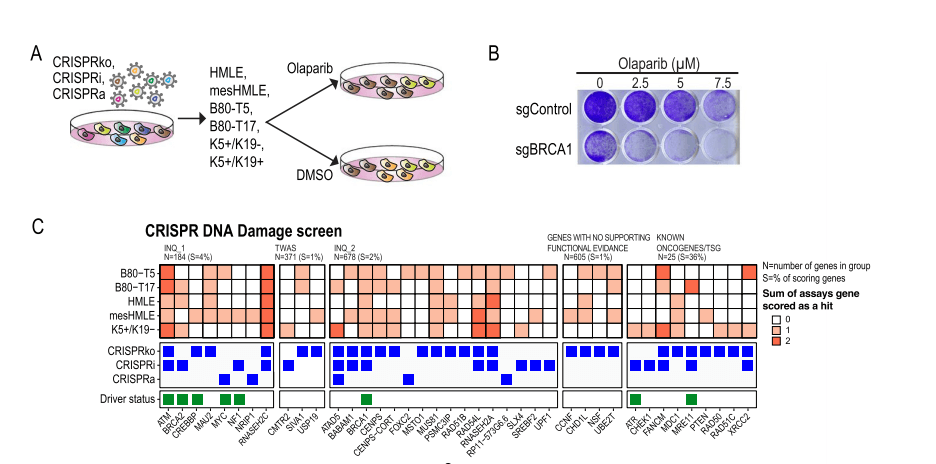
Figure 7: Olaparib Synthetic Lethality Screening Identified Breast Cancer Risk Genes that Regulate DNA Repair Pathways [7]
Conclusion
After reviewing the three application examples above, it is evident that CRISPR libraries demonstrate diversity and flexibility in breast cancer research. For instance, in the selection of target genes, not only the whole genome can be chosen for screening, but also large numbers of genes obtained from other methods. In terms of cell types, not only wild-type cells can be used, but also gene-edited cells to simulate specific gene mutations. Regarding screening methods, it can be done in vitro or in vivo, and even a combination of in vitro and in vivo screening strategies can be adopted to enhance the accuracy and effectiveness of the screening.
In summary, the CRISPR library technology provides personalized and flexible solutions for breast cancer research. Based on specific research goals, CRISPR libraries can be personalized, adjusted, and optimized to screen high-confidence targets, paving the way for new approaches to breast cancer treatment.
References
[1] https://www.who.int/zh/news-room/fact-sheets/detail/breast-cancer
[2] Nolan, E., Lindeman, G. J., & Visvader, J. E. (2023). Deciphering breast cancer: from biology to the clinic. Cell, 186(8), 1708–1728.
[3] Leon-Ferre, R. A., & Goetz, M. P. (2023). Advances in systemic therapies for triple negative breast cancer. BMJ (Clinical research ed.), 381, e071674.
[4] Geyer CE Jr, Garber JE, Gelber RD, et al. Overall survival in the OlympiA phase III trial of adjuvant olaparib in patients with germline pathogenic variants in BRCA1/2 and high-risk, early breast cancer. Ann Oncol. 2022;33(12):1250-1268.
[5] Chen, Bohong et al. Transmembrane nuclease NUMEN/ENDOD1 regulates DNA repair pathway choice at the nuclear periphery. Nature cell biology vol. 25,7 (2023): 1004-1016.
[6] Wang, Xiaoqing et al. “In vivo CRISPR screens identify the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer immunotherapy target.” Cell vol. 184,21 (2021): 5357-5374.e22.
[7] Tuano NK, Beesley J, Manning M, et al. CRISPR screens identify gene targets at breast cancer risk loci. Genome Biol. 2023;24(1):59. Published 2023 Mar 29.