Published on: November 18, 2024
Views:
--
Q&A - How to Maintain High Coverage During Cell Pool Passaging?

The basic principle of CRISPR library screening is as follows: After successfully constructing a screening-ready cell pool containing cells with various gene mutations, a specific selective pressure (such as drug treatment, bacterial or viral infection, etc.) is applied to enrich for mutant cells that are resistant to the pressure. Through NGS sequencing and bioinformatics analysis, the sgRNA information from the enriched cells is obtained to identify the target genes related to the phenotype of interest.
In CRISPR library screening experiments, constructing a high-quality screening-ready cell pool is key to identifying potential targets. However, due to the complex, costly, and time-consuming nature of the cell pool construction process, many researchers interested in target screening have been “excluded from the user list”. To lower the barrier for researchers, Ubigene, based on its proprietary CRISPR-iScreen™ platform, offers the in-stock products of CRISPR Library Screening-ready Cell Pools, allowing researchers to obtain the desired cell pool in a cost-effective, time-efficient manner and proceed with downstream screening experiments. Currently, our products have helped hundreds of research teams identify their desired targets!
Many customers still have doubts about whether our screening-ready cell pool products can be used for downstream screening experiments, particularly regarding questions like how to ensure sgRNA coverage and whether the cell pool should be passaged. Today, we will answer some of these questions!
Q1: Can the Library Cell Pools be Passaged? Does Passaging Affect sgRNA Coverage?
Studies show that when the initial representation depth of the infected library cells is low (e.g., 50x), sgRNA coverage will significantly decrease as the number of passages increases. However, if the initial representation depth is maintained (e.g., >500x), passaging the cells up to 8 passages (P8) does not cause a significant loss in sgRNA coverage.
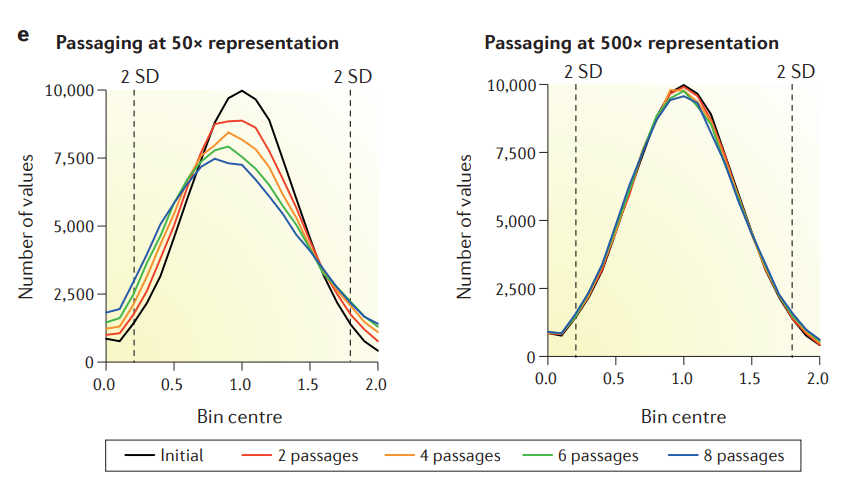
Figure 1. Effects of initial representative depth and passage number of library cell pools on sgRNA, respectively
Additionally, the likelihood of sgRNAs editing the genome increases with prolonged culturing. Some sgRNAs may only exhibit significant genome editing effects by day 14. Library cell pools at P0 may not have had sufficient time for most sgRNAs to generate efficient genome editing, which could result in the loss of cells that have not yet been edited, affecting experimental results.
To ensure that the majority of the cells in the library are edited and to ensure the reliability of downstream screening results, it is not recommended to use P0 cells directly for downstream drug screening experiments.
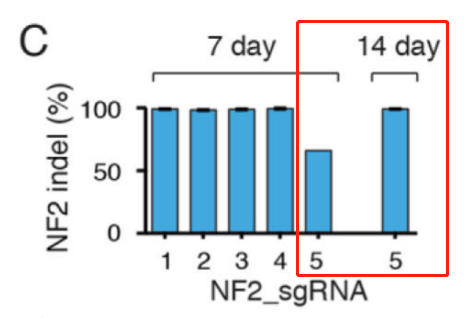
Figure 2. The editing efficiency of some sgRNAs in library cell pools is affected by the duration of cell culture
Q2: Does Freezing and Thawing Affect sgRNA Coverage? How is the Quality of the Library Cells Ensured?
Ubigene's CRISPR Library Screening-ready Cell Pools are infected and expanded at a high representation depth (>500x), and stored using high-quality serum-free cryopreservation medium (Product No.: YK-CR-100). The cells undergo cryopreservation, thawing, passaging, and next generation sequencing verification, no significant loss of sgRNA occurs after passaging.
As shown in the figure, we compared the sequencing results of different Ubigene’s whole-genome CRISPR Library Screening-ready Cell Pools at passages P0 and P5. After cryopreservation and thawing the P0 cells and expanding them to P5, the sgRNA coverage remained high, at over 99%!
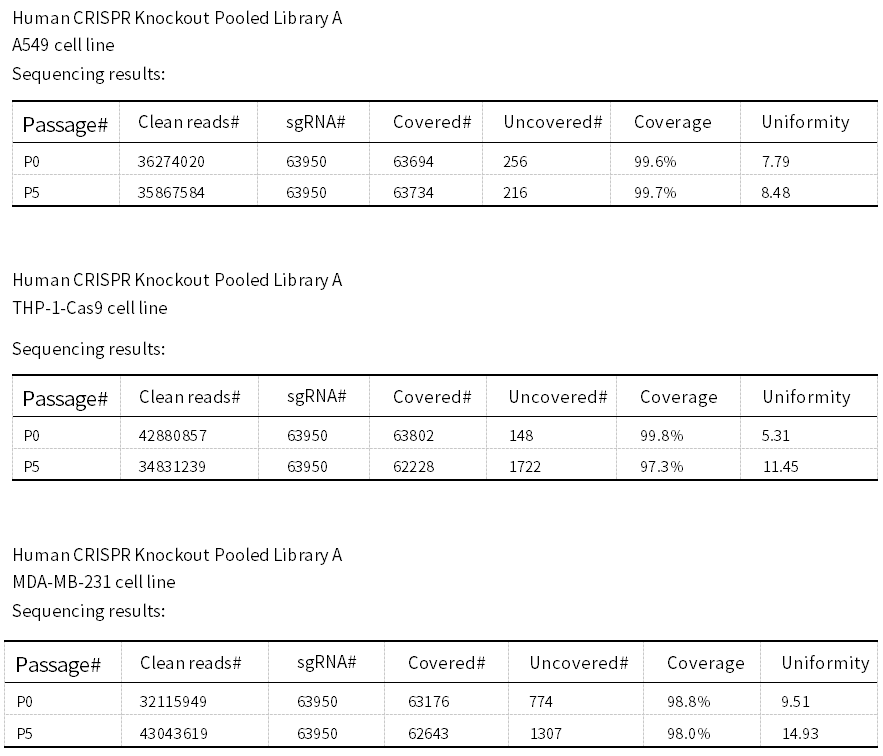
Figure 3. NGS results of different library cells at P0 and P5 passages
Q3: Can Ubigene's CRISPR Library Screening-ready Cell Pools be Used for Downstream Screening?
Ubigene's CRISPR Library Screening-ready Cell Pools with confirmed sgRNA coverage, can be used for downstream screening experiments with a reasonable extension of the culture time. Compared to P0 cells, these Cell Pools have a higher probability to be genome edited, making them suitable for downstream drug screening, flow cytometry sorting, viral infection, and other experiments.
It is worth mentioning that a small number of sgRNAs targeting essential genes may be lost during cell expansion due to their lethal effects on cells. While this does not affect the majority of library screening experiments, if the goal is to screen for cell-essential genes through passaging, this approach may not be suitable.
For experiments that require using cell passaging for library screening, it is recommended to use P0 cells and extend the passaging time of the library cells.
References
[1] Doench JG. Am I ready for CRISPR? A user's guide to genetic screens. Nat Rev Genet. 2018 Feb;19(2):67-80. doi: 10.1038/nrg.2017.97. Epub 2017 Dec 4.
[2] Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014 Jan 3;343(6166):84-87.










