Combination of HCT116 cells and CRISPR/Cas9: A perfect group for colorectal cancer treatment research
Cell information and applications
1. Cell information
HCT 116 was isolated from a 48 year old male patient with colorectal cancer in 1979 by M. brattain et al. The cells can form clones in semisolid agarose medium and are tumorigenic in athymic nude mice, forming epithelial-like tumors. So HCT 116 cell line is one of epithelial-like adherent tumor cell lines.
2. Cell applications and prospects
(1) To explore the mechanisms by which drugs affect colorectal cancer cell proliferation, migration and invasion. For example: PPARα plays an important role in the migratory activity and the expression of cyp2s1 and CYP1B1 of leukocyte treated HCT116 cells [2];
(2) To explore mechanisms of action in vitro by which drugs affect colorectal cancer cell growth, such as apoptosis and cell cycle alterations. For example: Raddeanin A regulates apoptosis and cycle arrest in human HCT116 cells through the PI3K/Akt pathway [3];
(3)To explore the sensitivity and resistance of drugs to colorectal cancer treatment. For example: Doxorubicin upregulates PD-L1 by inhibiting mir-140 expression in HCT116 cells, thereby rendering tumor cells being resistant to the drug [4];
(4)Development of new cancer-related signaling pathways to provide guidance for clinical treatment of colorectal cancer. For example, to investigate the significance of Notch and Wnt signaling pathways to drug resistance in colorectal cancer cell line HCT116, etc. [5];
(5)Developing new approaches of LncRNA and miRNAs in colorectal cancer treatment. For example: in HCT116 cells, long non-coding RNA of YWHAE competes with miR-323a-3p and miR-532-5p by activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways. It provides a new target site for colorectal cancer treatment. [6];
The application of CRISPR/Cas9 technology in HCT116
As mentioned above, HCT116 cells have been of great research value in the studies on various mechanisms of colon cancer, and the most basic research ideas generally start at the molecular level, that is, gene or protein. Thus, CRISPR/Cas9 technology has emerged in many research topics, and it is nonetheless an irreplaceable advantage in studying the functions of single carcinogenesis gene.
Some researches found that Trpm4 is highly expressed in human colorectal cancer and it seems to be associated with colorectal cancer cell proliferation, cell cycle and invasion. It was found that both tumor cell migration and invasion were indeed reduced by knocking out Trpm4 using CRISPR/Cas9 technology in HCT116 cells, also accompanied by cell cycle alterations [7];
As well as Cell reports that using CRISPR/Cas9 technology, replacing wild-type KRAS with mutant forms rendered the heterozygous mutant HCT116 cells more sensitive to drug treatment [8];
Moreover, by using CRISPR/Cas9 to knockout Tks4 in HCT116 cells, the cells exhibited significant epithelial mesenchymal transition, increased cell motility, and decreased cell-cell adhesion. Therefore found that the Tks4 gene plays an important role in EMT regulation and tumor development [9];
Researchers also found that the CTC1L1142H mutation resulted in impaired telomerase maintenance, by using CRISPR/Cas9 technology to mutate CTC1 in HCT116 cells. It confirmed that the CTC1:STN1 interaction was required for the inhibition of telomerase activity [10].
Therefore, there is no doubt among researchers for the favor of CRISPR/Cas9 technology. Ubigene, by providing the CRISPR/Cas9 technology services, has solved the experimental difficulties for many researchers, such as incomplete cell knockout, unstable cell knock-in, etc.. With our services, more and more researchers can achieve their scientific research objectives smoothly.
Case Studies
1. Poing mutation[11]
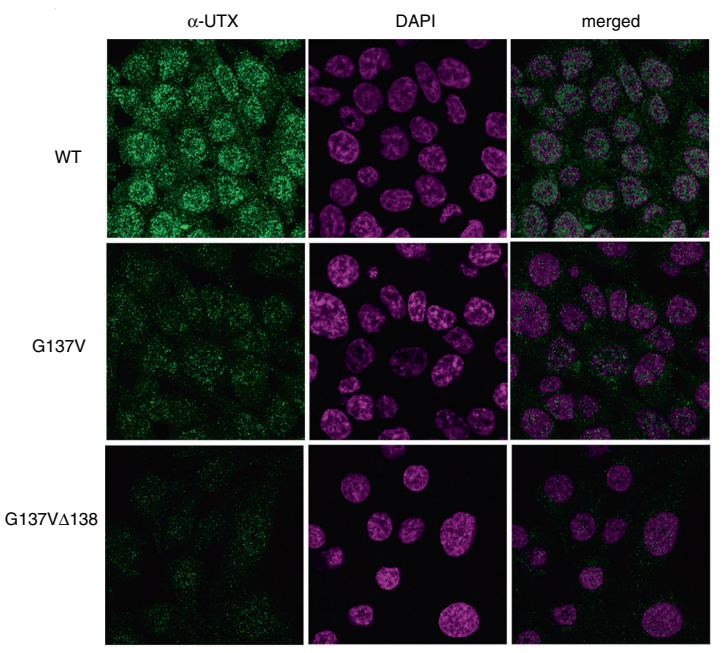
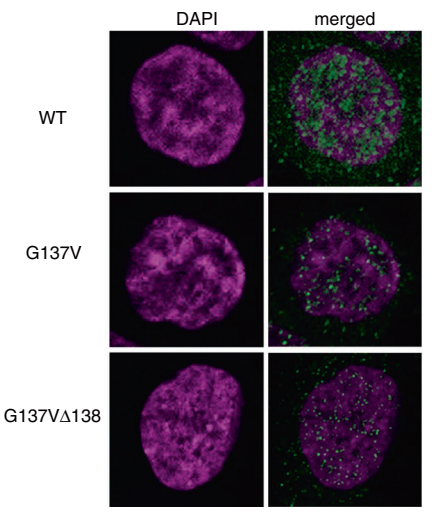
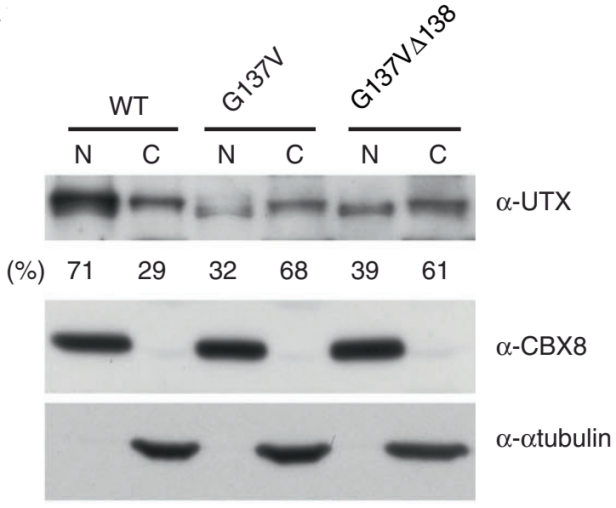
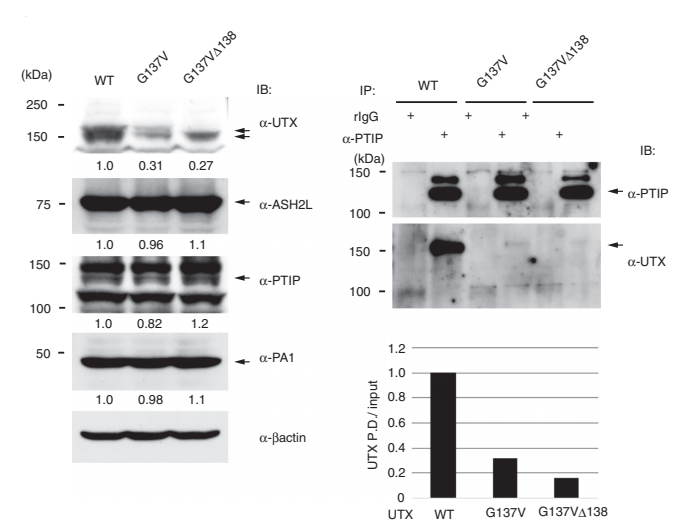
Hiroyuki Kato et al. using CRISPR/Cas9 technology introduced the mutations on UTX gene in HCT16 cells, which is G137V and G137VΔ138. They found that the wild-type UTX, which was originally expressed in the nucleus, greatly decreased in the nucleus, whereas increased in the cytoplasm in both mutant cells. As shown in Figure A to C: A, B were the immunofluorescence figures, C was the Western blot results. The results of co-immunoprecipitation are shown in fig.d, it is a novel regulatory mechanism of UTX. Also, the article also further revealed the importance of UTX interaction with MLL3/4 complex in cancer formation (ASH2L, PTIP and PA1 are components of MLL3/4 complex) .
2. Knockout [12]
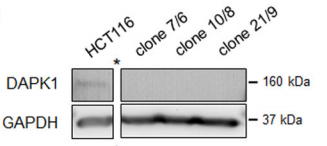
CRISPR/Cas9 gene-editing technology is used to obtain HCT116 cell line with DAPK1 deletion by Sara Steinmann et al, and finally revealed the effect of DAPK1 on the invasion of colorectal cancer.
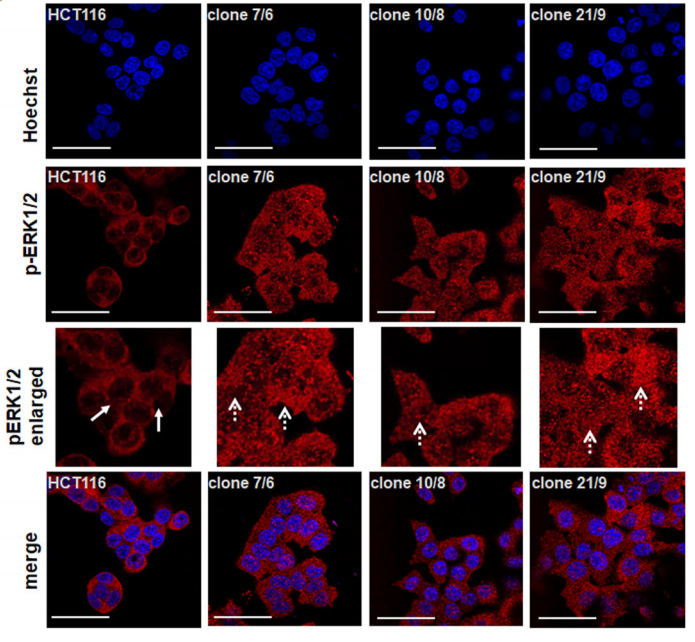
After Western Blot verification, three DAPK1 gene knockout monoclonal cells (Fig. A) were obtained by Sara Steinmann et al. Immunofluorescence assays detected pERK1/2 mainly in the wild-type HCT116 cytoplasm. However, in all three knockout cells, pERK1/2 was significantly expressed in the nucleus. (Fig. B)
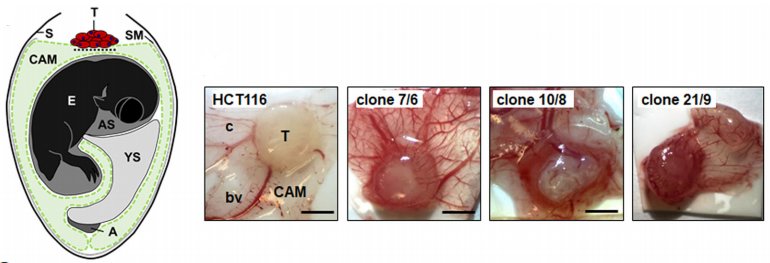
Because the chorionic allantoic membrane model (CAM) experiment is a classic in vivo model of angiogenesis, the researchers transplanted DAPK1 knockout clone and wild- type HCT116 cells to chicken CAM, and cultured in eggs for 5 days. It was found that the absence of DAPK1 led to the change of growth pattern and enhancement of tumor bud in CAM ( Fig. C)
In addition, the team used a rat brain 3D model in vitro to find that tumor cells had more proliferation in chicken embryonic organs, and the invasion ability was also enhanced. DAPK deficient HCT116 cells showed more diffuse tumor cells and preferentially accumulated in the liver, heart, and brain of the chicken embryo (Fig. D). Finally, the researchers found that DAPK1-ERK1 signaling pathway is involved in the metastasis of CRC (Fig. E).
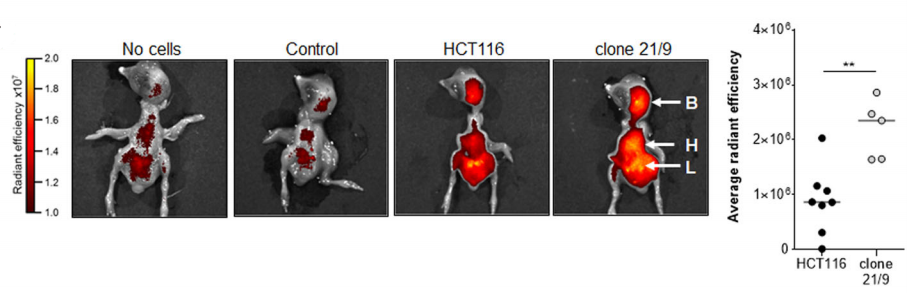
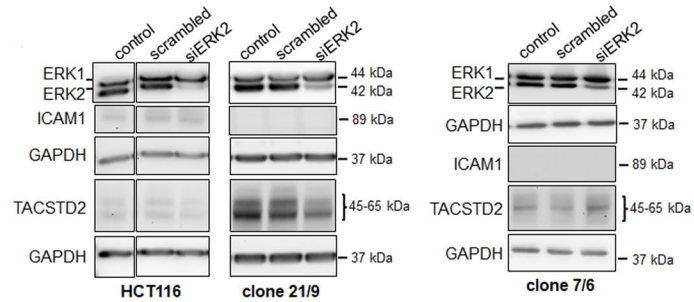
3. Knock-in [13]
BAX is one of the members of pro-apoptotic Bcl-2 gene family, which plays an important role in mitochondria dependent apoptosis initiation. R Peng et al have knocked in five mutation sites in BAX-KO HCT116 cells, which are located in the BAX gene and interact with other members of Bcl-2.
It has been reported that BAX-KO HCT116 cells do not undergo apoptosis by the stimulation of sulindac, a steroidal drug. R Peng et al found that knock in WT BAX in BAX-KO HCT116 cells can restore sulindac, and can restore HCT116 apoptosis caused by TRAIL. After knocking in the K21E and D33A mutations, BAX mediated apoptosis was completely restored; Knocking-in D68R and S184V mutations only partially recovered, while knocking in L70A / D71A mutation caused less apoptosis.
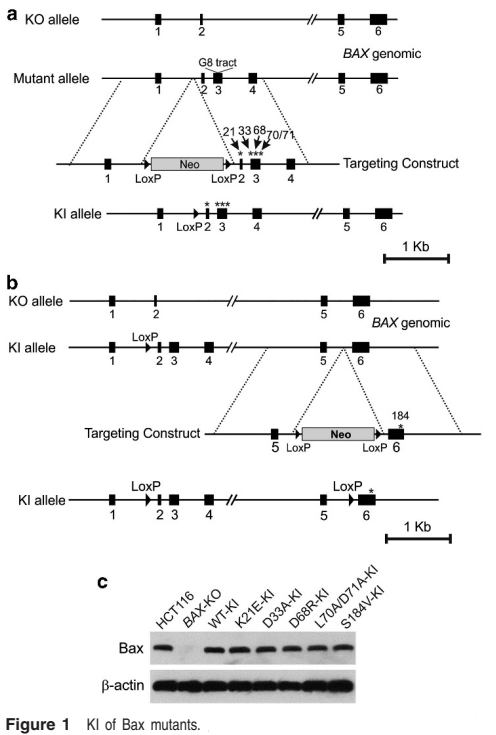
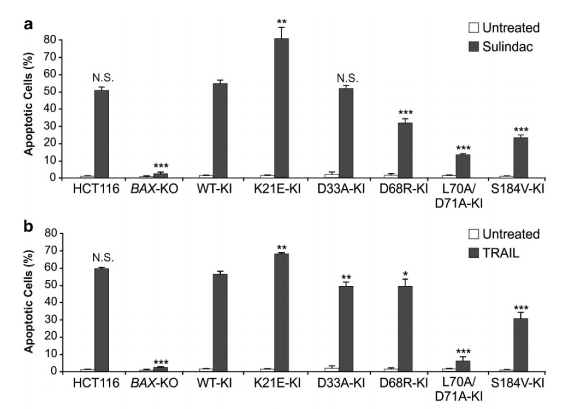
In this study, the target gene in the target cell is knocked out, and then knock in the target gene containing the mutation site and WT target gene (contrast), then we can clearly understand the specific site relationship between the protein expressed by the target gene and other proteins interacting with it, It is a good method to verify whether the predicted sites obtained by bioinformatics analysis really play a role in protein-protein interactionUbigene also provides the corresponding service convenience for scientific researchers. While enjoying the gene KI cell line service, free knockout cells can be obtained which meets the various research needs.
Reference:
[1]Bray, Freddie et al. “Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries.” CA: a cancer journal for cliniciansvol. 68,6 (2018): 394-424. doi:10.3322/caac.21492
[2]Khor CY, Khoo BY. PPARα plays an important role in the migration activity, and the expression of CYP2S1 and CYP1B1 in chrysin-treated HCT116 cells. Biotechnol Lett. 2020;42(8):1581-1595. doi:10.1007/s10529-020-02904-
[3]Meng C, Teng Y, Jiang X. Raddeanin A Induces Apoptosis and Cycle Arrest in Human HCT116 Cells through PI3K/AKT Pathway Regulation In Vitro and In Vivo. Evid Based Complement Alternat Med. 2019;2019:7457105. Published 2019 May 26. doi:10.1155/2019/7457105
[4]Naba NM, Tolay N, Erman B, Sayi Yazgan A. Doxorubicin inhibits miR-140 expression and upregulates PD-L1 expression in HCT116 cells, opposite to its effects on MDA-MB-231 cells. Turk J Biol. 2020;44(1):15-23. Published 2020 Feb 17. doi:10.3906/biy-1909-12
[5]Kukcinaviciute, Egle et al. “Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT116.” Journal of cellular biochemistry vol. 119,7 (2018): 5913-5920. doi:10.1002/jcb.26783
[6]Bjeije, Hassan et al. “YWHAE long non-coding RNA competes with miR-323a-3p and miR-532-5p through activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells.” Human molecular genetics vol. 28,19 (2019): 3219-3231. doi:10.1093/hmg/ddz146
[7]Kappel, Sven et al. “TRPM4 is highly expressed in human colorectal tumor buds and contributes to proliferation, cell cycle, and invasion of colorectal cancer cells.” Molecular oncology vol. 13,11 (2019): 2393-2405. doi:10.1002/1878-0261.12566
[8]Szeder, Bálint et al. “Absence of the Tks4 Scaffold Protein Induces Epithelial-Mesenchymal Transition-Like Changes in Human Colon Cancer Cells.” Cells vol. 8,11 1343. 29 Oct. 2019, doi:10.3390/cells8111343
[9]Burgess, Michael R et al. “KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer.” Cell vol. 168,5 (2017): 817-829.e15. doi:10.1016/j.cell.2017.01.020
[10]Gu, Peili et al. “CTC1-STN1 coordinates G- and C-strand synthesis to regulate telomere length.” Aging cell vol. 17,4 (2018): e12783. doi:10.1111/acel.12783
[11]Kato, Hiroyuki et al. “Cancer-derived UTX TPR mutations G137V and D336G impair interaction with MLL3/4 complexes and affect UTX subcellular localization.” Oncogene vol. 39,16 (2020): 3322-3335. doi:10.1038/s41388-020-1218-3
[12]Steinmann, Sara et al. “DAPK1 loss triggers tumor invasion in colorectal tumor cells.” Cell death & disease vol. 10,12 895. 26 Nov. 2019, doi:10.1038/s41419-019-2122-z
[13]Peng, R et al. “Targeting Bax interaction sites reveals that only homo-oligomerization sites are essential for its activation.” Cell death and differentiation vol. 20,5 (2013): 744-54. doi:10.1038/cdd.2013.4

















