Gene-editing RAW 264.7 cell line
helps to study inflammation and osteoclast formation
Murine macrophage cell line (RAW264.7) is considered to be one of the best models of macrophages, because it can carry out pinocytosis and phagocytosis, and is widely used in the study of inflammation, immunity, apoptosis and tumor. RAW264.7 cells can respond to stimulation in vitro, and then produce multinuclear cells with the characteristics of complete osteoclast differentiation. It is widely used in the study of bone diseases such as rheumatoid arthritis, osteoporosis, osteolysis, periodontitis, etc.
Application of RAW 264.7 cell line
RAW264.7 is a monocyte/macrophage-like cell line derived from the transformed cell line of Abelson leukemia virus in BALB/c. Raw 264.7 is one of the most popular in vitro models of osteoclasts and inflammation.
1. Osteoclast formation research:
RAW 264.7 has been proved to be easy to differentiate into osteoclasts under the induction of RANKL. Unlike primary osteoclast precursors, Raw 264.7 does not require the addition of macrophage colony-stimulating factor (M-CSF).
2. Inflammation research:
RAW264.7 is the most commonly used in vitro model for the selection of active compounds with anti-inflammatory and studying inflammation. RAW264.7 cells can mimic inflammatory reaction and release or upregulate a variety of inflammatory mediators, such as nitric oxide (NO), cyclooxygenase-2 (COX-2), tumor necrosis factor - α (TNF - α), interleukin-6 (IL-6) and so on.
CRISPR/cas9 mediated miRNA-155 knockout model helps to explore the inhibition of proinflammatory cytokines production in rheumatoid arthritis
It is reported that rheumatoid arthritis (RA) affects more than 21 million people worldwide. RA is an autoimmune inflammatory disease that affects joints. It is characterized by infiltration of macrophages and lymphocytes, proliferation of synovial fibroblasts, and eventual joint destruction. Macrophages play an important role in the pathogenesis of RA. The number of macrophages in RA inflammatory synovium is greater than that in normal joints, which is positively correlated with the severity of joint pain and inflammation. Many drugs have been approved for the treatment of rheumatoid arthritis, gene or cell therapy.
MicroRNA 155 (miR-155) is found in the gene BIC in mouse chromosome 16 and human chromosome 21. In clinical and experimental models, miR-155 is related to the pathogenesis of RA because it is upregulated in synovial membrane and synovial fluid macrophages in patients. miR-155 (KD) knockdown can inhibit the production of pro-inflammatory cytokines. The mechanism by which miR-155 participates in the formation of RA may be multifaceted. One of them is that miR-155 targets the 3 untranslated regions of Src homology-2, which contains inositol phospholipase 1 (SHIP1), a negative factor for inflammation. Therefore, elevated miR-155 in RA leads to a decrease in SHIP1 levels and then higher production of pro-inflammatory cytokines.
Researchers used CRISPR/Cas9 technology to create mutations in the endogenous miR-155 gene in RAW264.7, and obtained miR-155 genome knockout (GKO) clones. Further analysis showed that under LPS stimulation, miR-155 GKO clones expressed higher levels of SHIP1, but produced fewer pro-inflammatory cytokines.
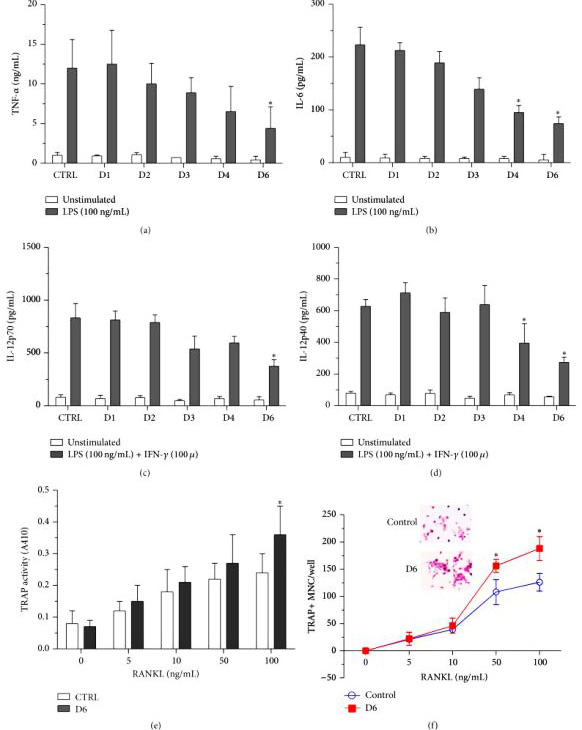
By using miR-155 GKO clones, removal of miR-155 resulted in decrease in proinflammatory cytokine production by macrophages, therefore confirming the previous observation that elevated miR-155 contributes to the sustained levels of cytokine production in RA patients. Researchers were able to reintroduce miR-155 effects by transfecting miR-155 mimics back into GKO clones. Altogether, these results indicate that the mutated endogenous miR-155 gene may cause the pre-miR-155 product to be truncated and fail to mature into a shorter but stable miR-155.
Knocking GFP into Raw264.7 by CRISPR/Cas9 to target NLRP3 inflammasome and exploring new targets for improving inflammatory diseases
The NLR family protein NLRP3 is a cytosolic sensor of exogenous pathogens and endogenous damage-associated molecular patterns (DAMPs). Upon activation, NLRP3 assembles with the adapter protein ASC and cysteine protease caspase-1 to form the NLRP3 inflammasome, resulting in the cleavage and activation of caspase-1. Activated capase-1 cleaves the precursors of IL-1β and IL-18 into mature forms and causes the release of several proinflammatory cytokines, including IL-1β and IL-18. It was reported that the NLRP3 inflammasome plays critical roles in the initiation and progression of diverse inflammatory diseases. Inhibition of the NLRP3 inflammasome signal has been shown to be effective in attenuating septic shock, peritonitis Alzheimer’s disease, atherosclerosis, T2D, multiple sclerosis and gout, among other diseases. Thus, the NLRP3 inflammasome is an excellent target for the treatment of multiple inflammatory diseases.
Using CRISPR/Cas9 to directly disrupt the key molecule-NLRP3 at the genomic level can not only completely inhibit the activation of NLRP3 inflammasome, but also avoid the potential risks of inhibiting off-target pathways of anti-inflammatory biologics and inhibitors. The development of a strategy to knockout NLRP3 with CRISPR/Cas9 is expected to be a more effective therapy for diverse inflammatory diseases.
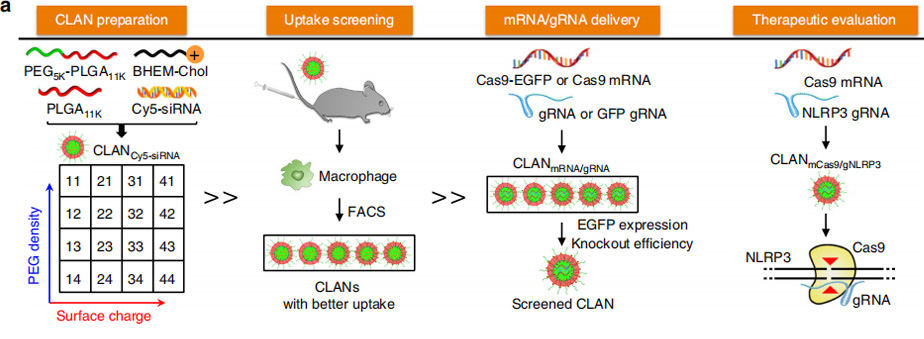
In this study, researchers reported a systemic delivery system of CRISPR/Cas9 by encapsulating mCas9 and gNLRP3 into CLAN. CLAN is a type of PEG-b-PLGA-based nanoparticle assisted by cationic lipid BHEM-Chol for nucleic acid therapeutics delivery. In their previous work, they have delivered small interfering RNA, RNA aptamers and hepatitis B virus CpG into tumor cells, cardiomyocytes, macrophages or plasmacytoid dendritic cells with CLAN.
However, mCas9/gNLRP3 are different from other nucleic acid therapeutics, and the properties of nanoparticles impact the efficiency of drug delivery. To test whether CLAN42 could effectively deliver mCas9/gRNA, Researchers encapsulated Cas9 and enhanced green fluorescent protein (EGFP) co-expressing mRNA (Cas9-EGFP mRNA, or mCas9-EGFP) and negative control gRNA (gNC) into selected CLANs (CLANmCas9-EGFP/gNC). Bone marrow-derived macrophages (BMDMs) were transfected with different CLANmCas9-EGFP/gNC. The CLAN42 transfection group showed the highest percentage of EGFP-positive BMDMs (65.8%). Next, they detected the gene knockout efficiency by transfecting Raw264.7 cells (a macrophage cell line) stably expressing GFP (Raw264.7-GFP) with CLANs encapsulating mCas9 and gRNA-targeting GFP (gGFP) (CLANmCas9/gGFP). The percentage of GFP-knockout (KO) Raw264.7-GFP cells in the CLAN42 transfection group was the highest and reached 53.9%. The in vivo mCas9/gRNA delivery efficiency of CLAN42 was further confirmed by injecting mice with different CLANmCas9-EGFP/gNC. The percentage of EGFP-positive peritoneal macrophages in the CLAN42 injection group was the highest (48.4%). Taken together, CLAN42 was the most effective CLAN in mCas9/gRNA delivery due to its highest ability of macrophage uptake, and CLAN42 was preferred to encapsulate mCas9/gNLRP3 (denoted as CLANmCas9/gNLRP3) for multiple inflammatory disease treatments.
Therefore, a library of CLANs of different surface charge and PEG density was created by adjusting the weight of the cationic lipid BHEM-Chol and mass fraction of PEG5K-b-PLGA11K in polymers. They screen CLANs both in vitro and in vivo and select a preferable CLAN to deliver mCas9/gNLRP3 into macrophages, which ameliorates LPS-induced septic shock, MSU-induced peritonitis and HFD-induced T2D by disrupting NLRP3 in macrophages. This study provides a promising strategy for the delivery of CRISPR/Cas9 into macrophages and the treatment of multiple inflammatory diseases.
This study confirmed that CLANmCas9/ gNLRP3 is a promising strategy for treating NLRP3-dependent inflammatory diseases and also provides an example for treating immune-related diseases by nanoparticles-mediated gene editing of immune cells.
Ubigene developed CRISPR-U™ (based on CRISPR/Cas9 technology) which is more efficient than general CRISPR/Cas9 in double-strand breaking, and CRISPR-U™ can greatly improve the efficiency of homologous recombination, easily achieve knockout (KO), point mutation (PM) and knockin (KI) in vitro and in vivo. With CRISPR-U™, Ubigene has successfully edit genes on more than 100 cell lines.
References
Jing W, Zhang X, Sun W, Hou X, Yao Z, Zhu Y. CRISPR/CAS9-Mediated Genome Editing of miRNA-155 Inhibits Proinflammatory Cytokine Production by RAW264.7 Cells. Biomed Res Int. 2015;2015:326042. doi:10.1155/2015/326042
Xu C, Lu Z, Luo Y, et al. Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases.[J]. Nature Communications, 2018, 9(1).
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6484317/








